Volume 29, Number 11—November 2023
Research
Genotypic Evolution of Klebsiella pneumoniae Sequence Type 512 during Ceftazidime/Avibactam, Meropenem/Vaborbactam, and Cefiderocol Treatment, Italy
Abstract
In February 2022, a critically ill patient colonized with a carbapenem-resistant K. pneumoniae producing KPC-3 and VIM-1 carbapenemases was hospitalized for SARS-CoV-2 in the intensive care unit of Policlinico Umberto I hospital in Rome, Italy. During 95 days of hospitalization, ceftazidime/avibactam, meropenem/vaborbactam, and cefiderocol were administered consecutively to treat 3 respiratory tract infections sustained by different bacterial agents. Those therapies altered the resistome of K. pneumoniae sequence type 512 colonizing or infecting the patient during the hospitalization period. In vivo evolution of the K. pneumoniae sequence type 512 resistome occurred through plasmid loss, outer membrane porin alteration, and a nonsense mutation in the cirA siderophore receptor gene, resulting in high levels of cefiderocol resistance. Cross-selection can occur between K. pneumoniae and treatments prescribed for other infective agents. K. pneumoniae can stably colonize a patient, and antimicrobial-selective pressure can promote progressive K. pneumoniae resistome evolution, indicating a substantial public health threat.
The number of deaths caused by antimicrobial resistance was estimated at 1.27 million worldwide in 2019 (1), mainly attributed to 6 bacterial species: Escherichia coli, Staphylococcus aureus, Klebsiella pneumoniae, Streptococcus pneumoniae, Acinetobacter baumannii, and Pseudomonas aeruginosa. To reinforce the antimicrobial drug pipeline, the European Union’s European Medicines Agency authorized the clinical use of ceftazidime/avibactam (CZA) in 2016, meropenem/vaborbactam (MVB) in 2018, and cefiderocol (FDC) in 2020. Although those drugs all belong to the β-lactams class, substantial differences in mechanisms of action and antimicrobial spectra exist among them. CZA is a third-generation cephalosporin (ceftazidime) combined with a diazabicyclooctane β-lactamase inhibitor (avibactam). Avibactam prevents class A (including K. pneumoniae carbapenemase [KPC]), class C, and some class D β-lactamases from hydrolyzing ceftazidime, restoring ceftazidime activity in KPC- and oxacillinase-48–producing Enterobacterales but not in metallo-β-lactamase producers (2). MVB is a carbapenem (meropenem) combined with a boronate β-lactamase inhibitor (vaborbactam); vaborbactam inhibits class A but not class D or B carbapenemases (3). FDC is a siderophore cephalosporin that has a catechol moiety on the C-3 side chain (4), which can form a chelating complex with ferric iron; thus, FDC is subject to active transport through the iron transport system, including TonB-dependent receptors (5). In addition, FDC is highly stable against β-lactamase activity (4,5).
In the past decade, Italy has seen a large increase in cases of carbapenem-resistant K. pneumoniae (1), mainly from 3 major KPC-producing (6) sequence types (STs): 101, 307, and 512 (7). In settings highly endemic for KPC-producing Enterobacterales, the selection of CZA-resistant, KPC-producing variants is of great concern (8,9). FDC resistance is not a 1-dimensional phenomenon (10); mutations in siderophore receptors (11), as well as in variants of β-lactamases KPC-2, CMY-2, CTX-M-15, and NDM-1, can confer FDC resistance (12). Nonetheless, cirA siderophore disruption substantially hinders bacterial fitness (13), and β-lactamase evolution contributing to FDC resistance typically comes at the price of functional trade-offs against other β-lactams (12). Under FDC treatment, in vivo resistance has been reported sporadically in Enterobacter cloacae (14) and Escherichia coli (15).
We describe a patient in Rome, Italy, who was colonized by carbapenem-resistant K. pneumoniae ST512. We integrated whole-genome sequencing, clinical, and microbiologic data to reconstruct the evolution of K. pneumoniae antimicrobial resistance in this patient after treatments for respiratory tract infections caused by different bacteria.
Case Report
In February 2022, a 62-year-old patient who was positive for SARS-CoV-2 was transferred from a long-term care facility to Policlinico Umberto I (PUI) in Rome. The patient was hospitalized for 95 days initially in the COVID-19 intensive care unit (ICU) and then in the general ICU until death, which was caused by gastrointestinal bleeding. The patient’s medical history was notable for bipolar disorder, obesity, inflammatory bowel disease, type 2 diabetes, respiratory failure, chronic kidney failure, and heart failure. Six months before transfer to PUI, the patient had a percutaneous endoscopic gastrostomy performed because of severe gastrointestinal bleeding caused by underlying inflammatory bowel disease. During the patient’s hospitalization at PUI, we isolated a total of 5 different K. pneumoniae strains from patient rectal swab samples and respiratory tract specimens.
Isolation of K. pneumoniae Strains and Susceptibility Testing
We identified the 5 K. pneumoniae strains by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (Bruker Daltonik GmbH, https://www.bruker.com). We identified carbapenemase genes by PCR using the GeneXpert system (Cepheid, https://www.cepheid.com) and used the lateral flow immunochromatography systems (NG-Test CARBA 5; Hardy Diagnostics, https://www.hardydiagnostics.com). We tested antimicrobial drug susceptibility by using the MicroScan WalkAway system (Beckman Coulter, Inc., https://beckman.com). We used gradient strips (Liofilchem, https://www.liofilchem.com) to test MVB and CZA MICs and the Compact Antimicrobial Susceptibility Panel (ComASP; Liofilchem) to test FDC MICs.
Whole-Genome Sequencing and Assembly
We performed whole-genome sequencing for each isolate. We purified genomic DNA by using the Isolate II Genomic DNA Kit (Bioline, https://www.bioline.com). We sent DNA to an external service to perform short-read Illumina sequencing (Illumina Inc., https://www.illumina.com). We obtained long reads by using the MinION Mk1C sequencing platform (Oxford Nanopore Technologies, https://nanoporetech.com). We extracted DNA for long reads by using the Wizard HMW DNA Extraction Kit (Promega, https://www.promega.com) and prepared libraries by using the Rapid Barcoding Kit 96; we sequenced libraries on R9.4.1 flow cells (Oxford Nanopore Technologies). We performed long-read assemblies by using Flye (16) with standard parameters. We integrated Illumina reads and Oxford Nanopore Technologies assemblies by using the Unicycler tool (17) in normal bridging mode and refined results by using the Bandage tool (18).
Genomic and Phylogenetic Analyses
We analyzed single-nucleotide polymorphisms (SNPs) among the 5 sequenced genomes in this study by using the Snippy tool (https://github.com/tseemann/snippy). We annotated the 5 genomes by using Rapid Annotation using Subsystem Technology and compared by using SEED software (19).
We used Prokka software (20) to annotate 133 genomes belonging to ST512: 5 sequences from this study, 12 from a previous study performed at PUI (8), and 116 downloaded from the Pasteur Institute BIGSdb database (https://bigsdb.pasteur.fr/klebsiella). We analyzed the resulting general feature formats by using Roary software (21) to build a core genome alignment and generated a consensus phylogenetic tree by using 1,000 ultrafast bootstraps (22) in IQ-TREE 2 (23) and the transversion model plus base frequency plus proportion of invariable sites nucleotide substitution model (24). We visualized tree and metadata by using Microreact (25) and adjusted those data by using open source InkScape software (https://www.inkscape.org). We assessed all genomes for replicons by using PlasmidFinder (26), for capsular polysaccharide and lipopolysaccharide loci by using Kaptive (27), and for virulence and resistance gene content by using Kleborate (28) software.
Cloning blaKPC-154 in pCR-Blunt II TOPO Vector
We cloned the novel β-lactamase blaKPC-154 allele from isolate 6099 into pCR-Blunt II TOPO-NeoR Vector and transformed TOP10 E. coli cells (both Thermo Fisher Scientific, https://www.thermofisher.com); we confirmed correctness of the cloned insert by Sanger sequencing. We tested the KPC-154 TOP10 E. coli clone for antimicrobial drug susceptibility by using the MicroScan system and measured CZA MICs by gradient tests (Liofilchem) as previously described.
OmpK36 Variant Modeling
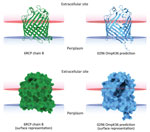
We predicted structures of the outer membrane porin (Omp) K36 from isolate 0296 in silico by using Alphafold2 on the European Galaxy server (https://usegalaxy.eu) and analyzed those structures by using UCSF ChimeraX (29) for both the cartoon (ribbon) and surface images. We compared the structures with chain B in the crystal structure of OmpK36 from a K. pneumoniae ST258 clinical isolate that has a GD amino acid insertion (30). We used the Orientations of Proteins in Membranes database (31) to obtain spatial arrangements of the protein structures in lipid bilayers.
Data Availability
We submitted the sequences of the strains analyzed in this study to GenBank. Whole-genome sequences are under Bioproject no. PRJNA992043 and complete plasmid sequences under accession nos. OQ096263 (pKpQIL-6099), OQ282880 (pIncA-6379), and OQ282881 (pIncA-0296).
Ethics Approval
According to the hospital’s routine practice, the patient or his relatives gave informed consent to share data for research purposes during hospital admission. The study protocol was approved by the Ethics Committee of Azienda Ospedaliero-Universitaria Policlinico Umberto I (approval no. 109/2020).
K. pneumoniae Strain Descriptions
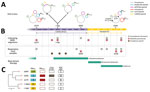
On the first day of the patient's hospitalization at PUI, surveillance rectal swab samples tested positive for K. pneumoniae (strain 6379) that produced both KPC and Verona integron-encoded metallo-β-lactamase (VIM) (Figure 1). The patient had been treated with CZA at the long-term care facility for a previous carbapenem-resistant K. pneumoniae bloodstream infection; treatment was discontinued upon admission to PUI.
After 17 days of hospitalization, a respiratory tract infection caused by Providencia stuartii (>100,000 CFU/mL in bronchoalveolar lavage fluid) developed in the patient, accompanied by pleural effusion, which we treated with CZA. After 1 week of treatment, we isolated 2 CZA-resistant, meropenem-susceptible K. pneumoniae strains from patient rectal swab samples but not respiratory tract specimens; both strains produced KPC but not VIM and had dimorphic colony phenotypes: white (strain 1186W) and transparent (strain 1186T). Because of potential pleural infection, we maintained CZA therapy for ≈40 days, resulting in complete eradication of P. stuartii from the respiratory tract.
On day 56, the patient was SARS-CoV-2 negative and was transferred to the general ICU at PUI. Respiratory tract samples were negative for P. stuartii but positive (>1,000 CFU/mL lavage fluid) for CZA-resistant K. pneumoniae (strain 6099) that produced KPC. CZA therapy was stopped, and MVB treatment was begun and continued until day 75. On day 75, respiratory tract samples tested negative for K. pneumoniae but positive for carbapenem-resistant A. baumannii (1,000 CFU/mL lavage fluid). Because of subsequent worsening respiratory conditions and chest imaging suggestive of new onset pneumonia, we replaced MVB therapy with FDC therapy (day 79) to treat A. baumannii pulmonary infection. On day 88, during FDC treatment, respiratory tract samples tested positive for both A. baumannii and K. pneumoniae (strain 0296) (1,000 CFU/mL lavage fluid each); strain 0296 was positive for KPC and VIM carbapenemases.
Phylogeny, Antimicrobial Resistance, and General Features of ST512
We assigned the 5 K. pneumoniae isolates from the patient (6379, 1186W, and 1186T from rectal swab samples, 6099 and 0296 from respiratory tract samples) to ST512 by using in silico multilocus sequence typing of whole-genome sequences. We aligned the complete genomes of those strains against 116 ST512 genomes retrieved from the Pasteur Institute K. pneumoniae BIGSdb database (December 7, 2022) and 12 ST512 genomes previously identified at PUI (8,32). Phylogeny of 4,654 core genes showed how isolates from the patient clustered together in the same clade with ST512 strains isolated at PUI during 2018–2020 (Figure 2) and were separated by 0–11 SNPs (Appendix 1 Table 1).
All ST512 isolates are defined by conserved features: the wzi allele 154 (assigned to the KL107 capsule), typical of the main clade II of clonal group 258 (33); O1/O2v2 O locus, serotype O2afg; 35Q mutation in the chromosome-encoded SHV-11 β-lactamase (28); premature stop at codon 89 in the OmpK35 protein, in most isolates coupled with a GD amino acid insertion in the OmpK36 eyelet (34); and chromosome mutations in the gyrA (S83I) and parC (S80I) genes potentially conferring quinolone resistance (35). The 5 isolates in this study had additional common features diverging from the average ST512 strain: a SNP in the mgrB gene, which stops translation at aa 29 of the MgrB regulator protein, conferring resistance to colistin (36); a SNP in the uhpB gene, which stops translation at aa 206 of the UhpB protein (37), conferring resistance to fosfomycin; and the presence of the locus encoding yersiniabactin siderophore, designated as YbST157 (38). The yersiniabactin siderophore is not common in ST512 isolates because this locus could only be found in 31/133 (23%) ST512 genomes from the BIGSdb collection.
The 5 K. pneumoniae strains carried multiple acquired resistance genes (Appendix 2 Table 1). Plasmid content of the 5 ST512 strains from the patient was not homogeneous: strains 6379 (first strain isolated) and 0296 (last strain isolated) both carried the pKpQIL plasmid containing the blaKPC-3 gene and IncA plasmid carrying the blaVIM-1 gene (Figures 1, 2). Strains 1186W, 1186T, and 6099 lacked the IncA-blaVIM-1 plasmid.
KPC Variants Conferring Resistance to Ceftazidime/Avibactam
On day 24, K. pneumoniae strains 1186W and 1186T colonized the patient (isolated from rectal swab samples). Those strains were negative for VIM-1 (loss of plasmid pIncA-blaVIM-1) but reached high levels of CZA resistance through the production of KPC-31. The blaKPC-31 gene replaced blaKPC-3 on an otherwise indistinguishable pKpQIL plasmid; the plasmid was integrated in the chromosome only in strain 1186W. The KPC-31 variant is characterized by a D179Y substitution in the Ω loop, previously shown to confer CZA resistance but restores carbapenem susceptibility (39).
On day 51, CZA-resistant K. pneumoniae strain 6099 caused a respiratory tract infection in the patient (Table). This strain hosted a pKpQIL plasmid encoding a novel KPC variant that was assigned by GenBank as KPC-154 (accession no. OQ096263) and was negative for the VIM-encoding IncA plasmid. Compared with KPC-3, KPC-154 has an RAPNKDDKYS amino acid duplication in position 263–273, corresponding to the 270 loop (40), but no differences are present in the Ω loop. When compared with an isogenic KPC-31–carrying strain, the strain carrying KPC-154 had higher MICs for several β-lactams (Appendix 2 Table 2).
Cefiderocol Resistance in ST512 Isolates Co-Producing KPC and VIM
K. pneumoniae strain 0296 (pulmonary infectious agent, isolated on day 88) was genotypically related to strain 6379 (colonizer, isolated on day 1). Both strains had similar plasmid content, including pKpQIL carrying blaKPC-3 and pIncA carrying blaVIM-1 (Figures 1, 2). However, isolate 0296 carried a pKpQIL-pKPN fused plasmid carrying catA1, dfrA12, aadA2, aph(3′)-Ia genes, 2 copies of mph(A), sul1, and ΔqacE genes (Appendix 2 Table 1), and the previously described putative gene involved in iron acquisition (fec) (41), all of which were not present in plasmid pKPN from isolate 6379. Within the core genome, strain 0296 had 7 SNPs and 3 deletions when compared with strain 6379 (Appendix 1 Table 2); 1 SNP created a premature stop codon at position 133 in the iron transporter protein CirA (Table; Figure 1) (42). Strain 0296 showed 4-fold higher FDC MIC than strain 6379 (6379 FDC MIC = 8 mg/L; 0296 FDC MIC = 32 mg/L). Moreover, in strain 0296, a conservative in-frame deletion resulted in a novel mutation within OmpK36; the deletion was 26 aa (residues Thr263 to Tyr289) according to the reference OmpK36 crystal structure in the Protein Data Bank (no. 6RCP; https://www.rcsb.org) (30). The in silico predicted 3-dimensional structure of the mutated OmpK36 porin showed a deep lateral cave, probably favoring MVB permeability from the extracellular site into the periplasmic space (Figure 3). Strain 0296 carrying the mutated OmpK36 protein showed a 5-fold reduction in MVB MICs compared with strain 6379 (6379 MVB MIC = 1.5 mg/L; 0296 MVB MIC = 0.047 mg/L).
We report the in vivo evolution of antimicrobial resistance in K. pneumoniae high-risk ST512 strains (43) that colonized or infected a critical patient during a 95-day hospitalization. From both phenotypic and genotypic perspectives, each K. pneumoniae isolate had univocal characteristics. The rare mutation in the UhpABC signaling pathway conferred fosfomycin resistance by altering expression of the hexose phosphate transporter (44,45). Mutations in the mgrB gene, identified in the 5 strains isolated in this study, 8 strains isolated during 2018–2020 at the PUI (8), and globally in 37/133 (27.8%) whole-genome sequences from the Pasteur Institute ST512 collection, conferred colistin resistance. Moreover, the 5 strains from this study carried the yersiniabactin locus, a virulence factor rarely reported in ST512 clones. The yersiniabactin locus was identified in a K. pneumoniae ST512 cluster sampled in the framework of the SPARK project from a single city located in northern Italy during 2017–2018 (46). All of those factors support the hypothesis that a single K. pneumoniae clone was obtained from the patient first from rectal swab samples and subsequently from the respiratory tract, rather than antimicrobial drug treatment selecting 5 distinct strains.
The development of β-lactam resistance in K. pneumoniae ST512 strains is a critical concern. At the time of transfer to PUI from a long-term care facility, the patient was colonized by a carbapenem-resistant K. pneumoniae strain that produced KPC-3 and VIM-1. Although KPC-3 production by K. pneumoniae is a persistent phenomenon in Italy (6), the spread of VIM-producing K. pneumoniae is less common. However, we previously described an Enterobacterales infection outbreak at PUI caused by IncA plasmids encoding VIM (47). Therefore, we hypothesize that an elusive and untraced spread of VIM-producing Enterobacterales might exist in hospitals and long-term care facilities in Rome.
During the first 3 weeks of hospitalization, the patient was not treated with β-lactams. We introduced CZA therapy to treat a respiratory tract infection sustained by P. stuartii, a pathogen more frequently associated with urinary tract infections (48) but also related to respiratory tract infections (49,50). After 3 days of CZA treatment, a KPC-31–producing, CZA-resistant K. pneumoniae strain colonized in the patient and then evolved to an infection by a KPC-154–producing CZA-resistant variant. The KPC-154 clone had a wild type Ω loop but a 13 aa insertion in the 270 loop of the enzyme, a feature previously described in other KPC variants (8).
We used MVB to treat the KPC-154–producing K. pneumoniae infection, which eradicated the K. pneumoniae strain from respiratory tract samples. However, a respiratory tract infection sustained by A. baumannii developed in the patient, who we then treated with FDC. Under FDC treatment, the respiratory tract sample tested positive for K. pneumoniae ST512 that was identical to the first colonizing strain, producing both KPC-3 and VIM-1 carbapenemases. This result suggests that a mixed population of KPC/VIM–positive and KPC-positive/VIM-negative K. pneumoniae was present in the patient during the entire hospitalization period. VIM-1–negative strains prevailed under CZA treatment, whereas MVB and FDC treatments selected the expansion of VIM-1–producing strains, which likely remained in a hidden reservoir within the patient.
The reemerged KPC/VIM producer had a high MIC for FDC because of the loss-of-function mutation in the CirA siderophore receptor. CirA mutations have previously been associated with FDC resistance in an in vitro model of New Delhi metallo-β-lactamase–producing K. pneumoniae (11) and in vivo for E. cloacae (14) and E. coli (15). However, the KPC/VIM–producing K. pneumoniae strain showed increased susceptibility to MVB, probably associated with a novel OmpK36 structure showing a large protein deletion; the 3-dimensional porin structure predicted a large lateral cave that might increase permeability of MVB. Nonetheless, the mutant OmpK36 could also be interpreted as a compensatory mutation that promotes survival of the CirA-defective strain.
We used last resort β-lactam–based antimicrobial drugs (CZA, MVB, and FDC) to treat the patient but only used MVB to treat the respiratory tract infection sustained by the CZA-resistant ST512 K. pneumoniae; other antimicrobial drugs were administered to treat infections caused by P. stuartii or A. baumannii. This case report serves as a warning that cross-selection can occur between K. pneumoniae and treatments prescribed against other infective agents and that K. pneumoniae can stably colonize a patient for a long period, promoting progressive evolution of the resistome under increasing selective pressure.
Dr. Arcari is a researcher at Sapienza University of Rome, Italy. His main interests focus on antimicrobial resistance mechanisms and their dissemination in bacterial pathogens.
Acknowledgment
This research was supported by European Union funding (to A.C., A.O., and G.A.) from the NextGeneration EU-MUR PNRR Extended Partnership Initiative on Emerging Infectious Diseases (project no. PE00000007, PE13 INF-ACT, Spoke 3).
References
- Murray CJL, Ikuta KS, Sharara F, Swetschinski L, Robles Aguilar G, Gray A, et al.; Antimicrobial Resistance Collaborators. Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet. 2022;399:629–55. DOIPubMedGoogle Scholar
- Shirley M. Ceftazidime-avibactam: a review in the treatment of serious gram-negative bacterial infections. Drugs. 2018;78:675–92. DOIPubMedGoogle Scholar
- Yahav D, Giske CG, Grāmatniece A, Abodakpi H, Tam VH, Leibovici L. New β-lactam-β-lactamase inhibitor combinations. Clin Microbiol Rev. 2020;34:e00115–20. DOIPubMedGoogle Scholar
- El-Lababidi RM, Rizk JG. Cefiderocol: a siderophore cephalosporin. Ann Pharmacother. 2020;54:1215–31. DOIPubMedGoogle Scholar
- Ito A, Nishikawa T, Matsumoto S, Yoshizawa H, Sato T, Nakamura R, et al. Siderophore cephalosporin cefiderocol utilizes ferric iron transporter systems for antibacterial activity against Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2016;60:7396–401. DOIPubMedGoogle Scholar
- David S, Reuter S, Harris SR, Glasner C, Feltwell T, Argimon S, et al.; EuSCAPE Working Group; ESGEM Study Group. Epidemic of carbapenem-resistant Klebsiella pneumoniae in Europe is driven by nosocomial spread. Nat Microbiol. 2019;4:1919–29. DOIPubMedGoogle Scholar
- Di Pilato V, Errico G, Monaco M, Giani T, Del Grosso M, Antonelli A, et al.; AR-ISS Laboratory Study Group on carbapenemase-producing Klebsiella pneumoniae. The changing epidemiology of carbapenemase-producing Klebsiella pneumoniae in Italy: toward polyclonal evolution with emergence of high-risk lineages. J Antimicrob Chemother. 2021;76:355–61. DOIPubMedGoogle Scholar
- Carattoli A, Arcari G, Bibbolino G, Sacco F, Tomolillo D, Di Lella FM, et al. Evolutionary trajectories toward ceftazidime-avibactam resistance in Klebsiella pneumoniae clinical isolates. Antimicrob Agents Chemother. 2021;65:
e0057421 . DOIPubMedGoogle Scholar - Hobson CA, Pierrat G, Tenaillon O, Bonacorsi S, Bercot B, Jaouen E, et al. Klebsiella pneumoniae carbapenemase variants resistant to ceftazidime-avibactam: an evolutionary overview. Antimicrob Agents Chemother. 2022;66:
e0044722 . DOIPubMedGoogle Scholar - Karakonstantis S, Rousaki M, Kritsotakis EI. Cefiderocol: systematic review of mechanisms of resistance, heteroresistance and in vivo emergence of resistance. Antibiotics (Basel). 2022;11:723. DOIPubMedGoogle Scholar
- McElheny CL, Fowler EL, Iovleva A, Shields RK, Doi Y. In vitro evolution of cefiderocol resistance in an NDM-producing Klebsiella pneumoniae due to functional loss of CirA. Microbiol Spectr. 2021;9:
e0177921 . DOIPubMedGoogle Scholar - Fröhlich C, Sørum V, Tokuriki N, Johnsen PJ, Samuelsen Ø. Evolution of β-lactamase-mediated cefiderocol resistance. J Antimicrob Chemother. 2022;77:2429–36. DOIPubMedGoogle Scholar
- Lan P, Lu Y, Jiang Y, Wu X, Yu Y, Zhou J. Catecholate siderophore receptor CirA impacts cefiderocol susceptibility in Klebsiella pneumoniae. Int J Antimicrob Agents. 2022;60:
106646 . DOIPubMedGoogle Scholar - Klein S, Boutin S, Kocer K, Fiedler MO, Störzinger D, Weigand MA, et al. Rapid development of cefiderocol resistance in carbapenem-resistant Enterobacter cloacae during therapy is associated with heterogeneous mutations in the catecholate siderophore receptor cirA. Clin Infect Dis. 2022;74:905–8. DOIPubMedGoogle Scholar
- Jousset AB, Poignon C, Yilmaz S, Bleibtreu A, Emeraud C, Girlich D, et al. Rapid selection of a cefiderocol-resistant Escherichia coli producing NDM-5 associated with a single amino acid substitution in the CirA siderophore receptor. J Antimicrob Chemother. 2023;78:1125–7. DOIPubMedGoogle Scholar
- Freire B, Ladra S, Parama JR. Memory-efficient assembly using Flye. IEEE/ACM Trans Comput Biol Bioinform. 2022;19:3564–77.
- Wick RR, Judd LM, Gorrie CL, Holt KE. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLOS Comput Biol. 2017;13:
e1005595 . DOIPubMedGoogle Scholar - Wick RR, Schultz MB, Zobel J, Holt KE. Bandage: interactive visualization of de novo genome assemblies. Bioinformatics. 2015;31:3350–2. DOIPubMedGoogle Scholar
- Overbeek R, Olson R, Pusch GD, Olsen GJ, Davis JJ, Disz T, et al. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Res. 2014;42(D1):D206–14. DOIPubMedGoogle Scholar
- Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–9. DOIPubMedGoogle Scholar
- Page AJ, Cummins CA, Hunt M, Wong VK, Reuter S, Holden MTG, et al. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics. 2015;31:3691–3. DOIPubMedGoogle Scholar
- Hoang DT, Chernomor O, von Haeseler A, Minh BQ, Vinh LS. UFBoot2: improving the ultrafast bootstrap approximation. Mol Biol Evol. 2018;35:518–22. DOIPubMedGoogle Scholar
- Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, von Haeseler A, et al. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol. 2020;37:1530–4. DOIPubMedGoogle Scholar
- Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 2017;14:587–9. DOIPubMedGoogle Scholar
- Argimón S, Abudahab K, Goater RJE, Fedosejev A, Bhai J, Glasner C, et al. Microreact: visualizing and sharing data for genomic epidemiology and phylogeography. Microb Genom. 2016;2:
e000093 . DOIPubMedGoogle Scholar - Carattoli A, Zankari E, García-Fernández A, Voldby Larsen M, Lund O, Villa L, et al. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother. 2014;58:3895–903. DOIPubMedGoogle Scholar
- Lam MMC, Wick RR, Judd LM, Holt KE, Wyres KL. Kaptive 2.0: updated capsule and lipopolysaccharide locus typing for the Klebsiella pneumoniae species complex. Microb Genom. 2022;8:
000800 . DOIPubMedGoogle Scholar - Lam MMC, Wick RR, Watts SC, Cerdeira LT, Wyres KL, Holt KE. A genomic surveillance framework and genotyping tool for Klebsiella pneumoniae and its related species complex. Nat Commun. 2021;12:4188. DOIPubMedGoogle Scholar
- Pettersen EF, Goddard TD, Huang CC, Meng EC, Couch GS, Croll TI, et al. UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci. 2021;30:70–82. DOIPubMedGoogle Scholar
- Wong JLC, Romano M, Kerry LE, Kwong HS, Low WW, Brett SJ, et al. OmpK36-mediated Carbapenem resistance attenuates ST258 Klebsiella pneumoniae in vivo. Nat Commun. 2019;10:3957. DOIPubMedGoogle Scholar
- Lomize MA, Lomize AL, Pogozheva ID, Mosberg HI. OPM: orientations of proteins in membranes database. Bioinformatics. 2006;22:623–5. DOIPubMedGoogle Scholar
- Jolley KA, Bray JE, Maiden MCJ. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res. 2018;3:124. DOIPubMedGoogle Scholar
- Deleo FR, Chen L, Porcella SF, Martens CA, Kobayashi SD, Porter AR, et al. Molecular dissection of the evolution of carbapenem-resistant multilocus sequence type 258 Klebsiella pneumoniae. Proc Natl Acad Sci U S A. 2014;111:4988–93. DOIPubMedGoogle Scholar
- García-Fernández A, Miriagou V, Papagiannitsis CC, Giordano A, Venditti M, Mancini C, et al. An ertapenem-resistant extended-spectrum-β-lactamase-producing Klebsiella pneumoniae clone carries a novel OmpK36 porin variant. Antimicrob Agents Chemother. 2010;54:4178–84. DOIPubMedGoogle Scholar
- Deguchi T, Fukuoka A, Yasuda M, Nakano M, Ozeki S, Kanematsu E, et al. Alterations in the GyrA subunit of DNA gyrase and the ParC subunit of topoisomerase IV in quinolone-resistant clinical isolates of Klebsiella pneumoniae. Antimicrob Agents Chemother. 1997;41:699–701. DOIPubMedGoogle Scholar
- Cannatelli A, Giani T, D’Andrea MM, Di Pilato V, Arena F, Conte V, et al.; COLGRIT Study Group. MgrB inactivation is a common mechanism of colistin resistance in KPC-producing Klebsiella pneumoniae of clinical origin. Antimicrob Agents Chemother. 2014;58:5696–703. DOIPubMedGoogle Scholar
- Verhamme DT, Arents JC, Postma PW, Crielaard W, Hellingwerf KJ. Glucose-6-phosphate-dependent phosphoryl flow through the Uhp two-component regulatory system. Microbiology (Reading). 2001;147:3345–52. DOIPubMedGoogle Scholar
- Lam MMC, Wick RR, Wyres KL, Gorrie CL, Judd LM, Jenney AWJ, et al. Genetic diversity, mobilisation and spread of the yersiniabactin-encoding mobile element ICEKp in Klebsiella pneumoniae populations. Microb Genom. 2018;4:
e000196 . DOIPubMedGoogle Scholar - Shields RK, Chen L, Cheng S, Chavda KD, Press EG, Snyder A, et al. Emergence of ceftazidime-avibactam resistance due to plasmid-borne blaKPC-3 mutations during treatment of carbapenem-resistant Klebsiella pneumoniae infections. Antimicrob Agents Chemother. 2017;61:e02097–16.PubMedGoogle Scholar
- Tooke CL, Hinchliffe P, Bonomo RA, Schofield CJ, Mulholland AJ, Spencer J. Natural variants modify Klebsiella pneumoniae carbapenemase (KPC) acyl-enzyme conformational dynamics to extend antibiotic resistance. J Biol Chem. 2021;296:
100126 . DOIPubMedGoogle Scholar - Villa L, Feudi C, Fortini D, Brisse S, Passet V, Bonura C, et al. Diversity, virulence, and antimicrobial resistance of the KPC-producing Klebsiella pneumoniae ST307 clone. Microb Genom. 2017;3:
e000110 . DOIPubMedGoogle Scholar - Zhang Z, Du W, Wang M, Li Y, Su S, Wu T, et al. Contribution of the colicin receptor CirA to biofilm formation, antibotic resistance, and pathogenicity of Salmonella Enteritidis. J Basic Microbiol. 2020;60:72–81. DOIPubMedGoogle Scholar
- Wyres KL, Holt KE. Klebsiella pneumoniae population genomics and antimicrobial-resistant clones. Trends Microbiol. 2016;24:944–56. DOIPubMedGoogle Scholar
- Zheng D, Bergen PJ, Landersdorfer CB, Hirsch EB. Differences in fosfomycin resistance mechanisms between Pseudomonas aeruginosa and Enterobacterales. Antimicrob Agents Chemother. 2022;66:
e0144621 . DOIPubMedGoogle Scholar - Ortiz-Padilla M, Portillo-Calderón I, de Gregorio-Iaria B, Blázquez J, Rodríguez-Baño J, Pascual A, et al. Interplay among different fosfomycin resistance mechanisms in Klebsiella pneumoniae. Antimicrob Agents Chemother. 2021;65:e01911–20. DOIPubMedGoogle Scholar
- Thorpe HA, Booton R, Kallonen T, Gibbon MJ, Couto N, Passet V, et al. A large-scale genomic snapshot of Klebsiella spp. isolates in Northern Italy reveals limited transmission between clinical and non-clinical settings. Nat Microbiol. 2022;7:2054–67. DOIPubMedGoogle Scholar
- Arcari G, Di Lella FM, Bibbolino G, Mengoni F, Beccaccioli M, Antonelli G, et al. A multispecies cluster of VIM-1 carbapenemase-producing Enterobacterales linked by a novel, highly conjugative, and broad-host-range IncA plasmid forebodes the reemergence of VIM-1. Antimicrob Agents Chemother. 2020;64:e02435–19. DOIPubMedGoogle Scholar
- Liu J, Wang R, Fang M. Clinical and drug resistance characteristics of Providencia stuartii infections in 76 patients. J Int Med Res. 2020;48:
300060520962296 . DOIPubMedGoogle Scholar - Molnár S, Flonta MMM, Almaş A, Buzea M, Licker M, Rus M, et al. Dissemination of NDM-1 carbapenemase-producer Providencia stuartii strains in Romanian hospitals: a multicentre study. J Hosp Infect. 2019;103:165–9. DOIPubMedGoogle Scholar
- Akbiyik A, Hepçivici Z, Eşer I, Uyar M, Çetin P. The effect of oropharyngeal aspiration before position change on reducing the incidence of ventilator- associated pneumonia. Eur J Clin Microbiol Infect Dis. 2021;40:615–22. DOIPubMedGoogle Scholar
Figures
Table
Cite This ArticleOriginal Publication Date: October 03, 2023
Table of Contents – Volume 29, Number 11—November 2023
| EID Search Options |
|---|
|
|
|
|
|
|

Please use the form below to submit correspondence to the authors or contact them at the following address:
Alessandra Carattoli, Department of Molecular Medicine, Sapienza University of Rome, Viale di Porta Tiburtina 28, Rome 00185, Italy
Top