Volume 29, Number 12—December 2023
Research Letter
Genome-Based Characterization of Listeria monocytogenes, Costa Rica
Abstract
Genomic data on the foodborne pathogen Listeria monocytogenes from Central America are scarce. We analyzed 92 isolates collected during 2009–2019 from different regions in Costa Rica, compared those to publicly available genomes, and identified unrecognized outbreaks. Our findings suggest mandatory reporting of listeriosis in Costa Rica would improve pathogen surveillance.
Listeria monocytogenes is a gram-positive pathogen responsible for listeriosis, a severe foodborne infection that causes high hospitalization and mortality rates in at-risk populations, including older adults, immunocompromised persons, pregnant women, and newborns (1). L. monocytogenes diversity can be classified into lineages, genoserogroups, clonal complexes (CCs), and sequence types (STs), defined by multilocus sequence typing (MLST) (2). Core-genome MLST (cgMLST) further identifies sublineages (SLs) and cgMLST types (CTs) (2). Major CCs and SLs are distributed globally and can be heterogeneous in terms of virulence; isolates from serogroup IVb (lineage I) often cause the most severe infections (2–4).
Pathogen surveillance using whole-genome sequencing (WGS) provides unprecedented resolution for identifying case clusters and contamination sources and for predicting strain virulence and antimicrobial resistance, which can aid in risk assessment (2,5). Previous studies confirmed L. monocytogenes in various foods in Costa Rica; reported contamination levels were 5%–20% in processed meat products and fresh cheeses (6,7). Because listeriosis is not a notifiable disease in Costa Rica, its prevalence is unknown, and diversity of L. monocytogenes circulating in the country is unclear.
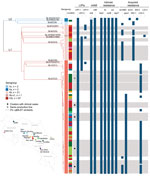
To clarify the diversity of and potential public health risk from circulating strains, we used WGS to characterize 92 isolates recovered during 2009–2019 from 16 clinical, 67 food, and 9 production environment samples in Costa Rica (Appendix). When location data were available, isolates were from urban areas, including the capital city San José, and from rural areas where fresh cheese production is prevalent, including Alajuela, Naranjo, San Ramón, Vara Blanca, Upala, and Turrialba. Turrialba region accounts for 70% of fresh cheese produced in Costa Rica (Figure; Appendix).
We found that isolates from lineage I (95%, n = 88) and lineage II (5%, n = 4) were unevenly distributed into 12 different SLs and CCs (Figure; Appendix Figure 1). Those isolates included a new lineage I sublineage, designated SL1079 (new MLST singleton ST1079), which was identified in an isolate from shrimp (cgMLST type L1-SL1079-ST1079-CT1669). That isolate had an atypical genoserogroup IIb profile, designated IIb-v1, that differed from the classic IIb profile by the presence of lmo0737. WGS confirmed the presence of lmo0737 and flanking genes lmo0733–39, typically found in lineage II isolates from serogroups IIa and IIc but only occasionally found in lineage I serogroup IVb-v1 (8). Of note, 80% of isolates investigated from both clinical and food-associated sources were from sublineages SL2/CC2 (66%, n = 61) and SL3/CC3 (14%, n = 13). SL2/CC2 (serogroup IVb) and SL3/CC3 (serogroup IIb) isolates are found worldwide and are associated with invasive infections (2–4). However, they are rarely the most prevalent genotypes (2,3). Available data from other countries in Central America confirmed overrepresentation of SL2/CC2 and SL3/CC3 in Costa Rica (Appendix), which could be related to country’s geographic location, climatic peculiarities, commercial trends, or natural reservoirs.
At the strain level, we identified 48 CTs, of which 44 (92%) were not previouly reported. Eleven (23%) CTs included multiple isolates at a cutoff of 7 allelic differences of 1,748 cgMLST loci (2) (Table; Figure; Appendix Figures 1–3). Eight isolates were cgMLST type L1-SL2-ST2-CT2715, which accounted for 25% of clinical cases and spanned 9 years (Table).
Most human cases were associated with dairy products (Table). However, tracing to confirm the source of infection was not possible because most production is conducted by local farmers, often without traceability or attribution to the site of production.
Fresh cheese production is an economic staple in Costa Rica, and previous studies have reported L. monocytogenes detection in those products (7). Results from this study also show detection of identical strains of cgMLST type L1-SL2-ST2-CT6072 along the same production line, from raw materials to the final product, suggesting inadequate sanitation contributes to contamination (9).
L. monocytogenes is problematic for the food industry because it can survive and multiply under adverse environmental conditions (10). In this study, 90% of isolates carried >1 genetic element encoding for tolerance to disinfectants or stress. Markers of tolerance to disinfectants included qacA (51%, n = 47), bcrABC (23%, n = 21), and emrC (1%, n = 1). In addition, isolates had stress survival islet (SSI) genes, including SSI-1, conveying tolerance to low pH and high salt concentrations (21%, n = 19), and SSI-2 conveying, tolerance to high pH and oxidative stress (1%, n = 1), as well as Listeria genomic island (LGI) genes, including LGI-2 (50%, n = 48) and LGI-3 (1%, n = 1) conveying tolerance to metals. Those tolerances can make L. monocytogenes elimination from production sites more difficult.
This study provides insight into the diversity of L. monocytogenes strains circulating in Central America and can aid national reference institutions in promoting regulatory changes to guarantee mandatory listeriosis reporting. In addition, institutions should establish mechanisms to provide low-cost microbiologic analysis. We also recommend regular sampling of risk products and training of artisanal processors.
In conclusion, strengthened WGS surveillance in Costa Rica could assist in controlling L. monocytogenes and provide food producers with information on strain diversity and effective means of eradication. WGS surveillance also would enable authorities to detect outbreaks and trace sources of contamination.
Ms. Giralt-Zúñiga is currently a PhD student at the Molecular Microbiology Department of the Institute for Biology, Humboldt-Universität zu Berlin, Germany. Her research interests focus on infectious diseases and enteric pathogens.
Acknowledgments
We thank the submitters for depositing their data in public databases and Institut Pasteur teams for the curation and maintenance of Bacterial Isolate Genome Sequence Databases (BIGSdb) at Institut Pasteur, Paris, France (https://bigsdb.pasteur.fr). We also thank the CONAGEBIO from the Costa Rican Ministry of Environment and Energy (MINAE) for providing the permits to the access to biological material presented here (file no. CM-ITCR-003-2021).
This work includes multilocus sequence typing profiles publicly available on BIGSdb-Listeria (https://bigsdb.pasteur.fr/listeria).
This work was developed within the framework of agreements between Institut Pasteur and the University of Costa Rica, and between Institut Pasteur and Instituto Tecnológico de Costa Rica (amendment no.1 to the memorandum of understanding dated February 21, 2018), and supported by Institut Pasteur, Inserm, Santé Publique France, the government of France Investissement d’Avenir program Laboratoire d’Excellence Integrative Biology of Emerging Infectious Diseases (no. ANR-10-LABX-62-IBEID), the Vice Rectory of Research of the Instituto Tecnológico de Costa Rica (research project no. 1510160), and the Vice Rectory of Research at the University of Costa Rica (project nos. B2104 and B9026). Travel funds for M.G.Z. were provided by the Instituto Tecnológico de Costa Rica and the Consejo Nacional para Investigaciones Científicas y Tecnológicas (CONICIT). Travel funds for M.R.S. were provided by the Institut Français.
This article was preprinted at https://doi.org/10.1101/2023.06.23.543262.
References
- Swaminathan B, Gerner-Smidt P. The epidemiology of human listeriosis. Microbes Infect. 2007;9:1236–43. DOIPubMedGoogle Scholar
- Moura A, Criscuolo A, Pouseele H, Maury MM, Leclercq A, Tarr C, et al. Whole genome-based population biology and epidemiological surveillance of Listeria monocytogenes. Nat Microbiol. 2016;2:16185. DOIPubMedGoogle Scholar
- Chenal-Francisque V, Lopez J, Cantinelli T, Caro V, Tran C, Leclercq A, et al. Worldwide distribution of major clones of Listeria monocytogenes. Emerg Infect Dis. 2011;17:1110–2. DOIPubMedGoogle Scholar
- Maury MM, Tsai YH, Charlier C, Touchon M, Chenal-Francisque V, Leclercq A, et al. Uncovering Listeria monocytogenes hypervirulence by harnessing its biodiversity. Nat Genet. 2016;48:308–13. DOIPubMedGoogle Scholar
- Schmid D, Allerberger F, Huhulescu S, Pietzka A, Amar C, Kleta S, et al. Whole genome sequencing as a tool to investigate a cluster of seven cases of listeriosis in Austria and Germany, 2011-2013. Clin Microbiol Infect. 2014;20:431–6. DOIPubMedGoogle Scholar
- Calvo-Arrieta K, Matamoros-Montoya K, Arias-Echandi ML, Huete-Soto A, Redondo-Solano M. Presence of Listeria monocytogenes in ready-to-eat meat products sold at retail stores in Costa Rica and analysis of contributing factors. J Food Prot. 2021;84:1729–40. DOIPubMedGoogle Scholar
- Posada-Izquierdo GD, Mazón-Villegas B, Redondo-Solano M, Huete-Soto A, Víquez-Barrantes D, Valero A, et al. Modelling the effect of salt concentration on the fate of Listeria monocytogenes isolated from Costa Rican fresh cheeses. Foods. 2021;10:1722. DOIPubMedGoogle Scholar
- Leclercq A, Chenal-Francisque V, Dieye H, Cantinelli T, Drali R, Brisse S, et al. Characterization of the novel Listeria monocytogenes PCR serogrouping profile IVb-v1. Int J Food Microbiol. 2011;147:74–7. DOIPubMedGoogle Scholar
- Castro H, Jaakkonen A, Hakkinen M, Korkeala H, Lindström M. Occurrence, persistence, and contamination routes of Listeria monocytogenes genotypes on three Finnish dairy cattle farms: a longitudinal study. Appl Environ Microbiol. 2018;84:e02000–17. DOIPubMedGoogle Scholar
- Buchanan RL, Gorris LGM, Hayman MM, Jackson TC, Whiting RC. A review of Listeria monocytogenes: an update on outbreaks, virulence, dose-response, ecology, and risk assessments. Food Control. 2017;75:1–13. DOIGoogle Scholar
Figure
Table
Cite This ArticleOriginal Publication Date: November 14, 2023
1These first authors contributed equally to this article.
2Current affiliation: Humboldt-Universität zu Berlin, Berlin, Germany.
3Current affiliation: Universidad Autónoma de Chile, Temuco, Chile.
Table of Contents – Volume 29, Number 12—December 2023
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
Address for correspondence. Marc Lecuit or Javier Pizarro-Cerda, 28 Rue du Docteur Roux, 75724 Paris CEDEX 15, France
Top