Volume 30, Number 4—April 2024
Dispatch
Uncommon Salmonella Infantis Variants with Incomplete Antigenic Formula in the Poultry Food Chain, Italy
Abstract
Uncommon Salmonella Infantis variants displaying only flagellar antigens phenotypically showed identical incomplete antigenic formula but differed by molecular serotyping. Although most formed rough colonies, all shared antimicrobial resistances and the presence of usg gene with wild-type Salmonella Infantis. Moreover, they were undistinguishable wild-type Salmonella Infantis by whole-genome sequencing.
The emergence of variants posing threats to human health and animal production characterizes the epidemiology of Salmonella (1) and also S. enterica serovar Infantis (antigenic formula 6,7:r:1,5). Over the past few decades, this serovar has rapidly emerged along the poultry chain; as of 2023, it is the most prevalent serovar isolated from broiler chickens in the European Union and among the 4 most common serovars in humans (2,3). The European Commission identified Salmonella Infantis as a target serovar for which control measures must be implemented if it is isolated in breeding flocks of Gallus gallus chickens (4). Some of the mandatory control measures implemented in Italy are appropriate health measures applied at farms; eradication or slaughtering of Salmonella-positive birds and management of their carcasses in accordance with EC regulation no. 1069/2009; and disposal of eggs produced by Salmonella Infantis‒positive groups and additional cleaning and disinfection procedures of the flock environment and facilities. The strict and targeted control measures implemented in case of identification of Salmonella Infantis‒positive flocks (5) require standardized analytical methods (i.e., ISO 6579:1 for isolation of Salmonella and methods based on the Kaufmann-White scheme for serotyping or validated alternative methods) to ensure high quality of surveillance, prompt identification of positive samples, and prompt implementation of eradication measures.
Isolates with an incomplete antigenic formula that carried flagellar antigens typical of Salmonella Infantis (r:1,5), but lacked the somatic ones (6,7), have been increasingly isolated from poultry sources in Italy. Laboratories in charge of Salmonella controls and surveillance needed to quickly identify and characterize these isolates to ascertain if these atypical variants were Salmonella Infantis strains and consequently manage their isolation on farms for breeding G. gallus fowl. Our investigation also included strains showing those atypical features isolated from food matrices, to estimate their spread along the poultry chain.
We tested a total of 31 Salmonella strains that could be ascribed to Salmonella Infantis but lacked the expression of the complete antigenic formula from animals (N = 20) and food (N = 11). Those strains were isolated during 2014‒2022 from samples taken in different regions of Italy (Table). We included 1 wild-type Salmonella Infantis strain isolated from food as a reference. We serotyped all Salmonella isolates by slide agglutination with Salmonella antiserum samples (Statens Serum Institut, Copenhagen, Denmark); we assigned antigenic formulas in accordance with ISO 6579:3. If traditional serotyping could not provide conclusive results because the complete antigenic formula was not expressed, we performed molecular serotyping by using an xMAP Salmonella serotyping assay (6). In particular, a positive match from the somatic antigen with C1 probe (wzy gene) is expected in case of wild-type Salmonella Infantis isolates. Colony morphology was investigated on agar tryptose solid medium. We assessed antimicrobial susceptibility by MIC by using the broth microdilution method with the Sensititer EUVSEC panel (TREK Diagnostics System; ThermoFisher, https://www.thermofisher.com) and interpreted results in accordance with European Committee on Antibiotic Susceptibility Testing (EUCAST) epidemiologic cutoff values. We selected the usg gene (SIN_02055) as a marker gene specific for Salmonella Infantis; we used the PCR targeting this gene described by Yang et al. (7) as an identification marker to test whether strains belonged to Salmonella Infantis serovar. Finally, we performed whole-genome sequencing as described in Petrin et al. (8) and used a core genome multilocus sequence typing (cgMLST) scheme approach (9) to assess genetic relatedness among the investigated strains and with the wild-type Salmonella Infantis strain. We also performed in-silico serotyping as described by Costa et al. (9).
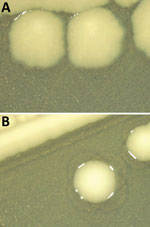
Thirty-one of 32 strains did not agglutinate with the somatic serum specimens for 6,7 antigens, indicating that their antigenic formula was -:r:1,5 (Table). Molecular serotyping identified a match with the C1 somatic probe in 9 strains. Of those, 3 were isolated from food and 6 from animal sources. For the remaining 22 strains, detection with the C1 somatic probe was negative. The detection of typical Salmonella Infantis flagellar antigens was positive for all the tested strains. In silico serotyping did not provide conclusive results on the O-antigen encoding region for 27 isolates; those results support the need for further analyses. Twenty-six strains displayed rough colony morphology (Figure 1), which in many cases contributes to increased sensitivity to immune defense, even if it remains unclear whether these strains are pathogenic (10). Most of the selected strains shared a similar resistance pattern to multiple antimicrobial drugs (ampicillin, ciprofloxacin, azithromycin, nalidixic acid, tetracycline, trimethoprim, and sulfamethoxazole) (Appendix Table) typical of the circulating wild-type Salmonella Infantis strains spread in broiler populations (1).
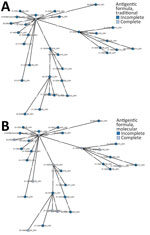
All the 31 isolates harboring incomplete antigenic formula, as well as the isolate with the complete antigenic formula, had usg gene present in their genomes; the presence of the gene can be considered a promising tool for rapid diagnosis of those atypical strains with incomplete expression of somatic antigens. The investigations carried out by cgMLST to establish the genetic relatedness among isolates did not enable us to identify different Salmonella Infantis populations on the basis of their antigenic formula, considering both traditional serotyping (Figure 2, panel A) and molecular serotyping (Figure 2, panel B).
Our results describe heterogeneous Salmonella Infantis strains widespread in the poultry food chain in Italy. This heterogeneity seems to involve the antigen-coding genetic context that probably would also affect the colony morphology (11). It is well known that rough morphology of Salmonella isolates originates from deletion or truncations of lipopolysaccharide O-antigen encoding genes (12). In our study, however, we could not link the rough phenotype, identified for the atypical strains, to the absence of the wzy gene nor to the isolation matrix and source. Further analyses at the genomic level will clarify the deletion/truncation pattern and identify the involved genes; the genetic region encoding for the O-antigen is composed of many different genes (13,14).
We found atypical strains that lacked the expression of the complete antigenic formula to be genetically indistinguishable from wild-type Salmonella Infantis. We also found a clear indication toward an antimicrobial resistance profile, shared by all the investigated atypical isolates, that provided resistance to multiple antimicrobial compounds commonly described for the wild-type circulating clones of Salmonella Infantis (1,15). This similarity leads to a likely closeness of those somatic variant isolates to the wild-type Salmonella Infantis isolates, showing a complete antigenic formula. The close relationships between Salmonella Infantis and variant isolates with an incomplete antigenic formula and the reasons for the lack of O-antigen expression should be further investigated. Identifying the atypical strains would pose a diagnostic issue because they would not be recognized as Salmonella Infantis by traditional serotyping, which remains by far the most common method used by laboratories in charge of Salmonella surveillance. This difficulty in identifying such atypical strains could compromise the prompt recognition of infected poultry flocks and hamper the timely implementation of the targeted control measures required by legislation, which would have serious repercussions on public health.
Dr. Petrin is a biotechnologist in the microbiology department of the National and WOAH Reference Laboratory for Salmonella in Italy. Her primary research interests are molecular epidemiology and antimicrobial drug resistance of Salmonella.
Acknowledgments
We thank Mario D’Incau, Elisabetta Di Giannatale, Elisa Goffredo, Maria Laura De Marchis, Alessia Zicavo, and Stefano Lollai, who contributed Salmonella strains for this study.
Raw sequence data were submitted to the European Nucleotide Archive (https://www.ebi.ac.uk/ena) under accession no. PRJEB72047.
This work was supported by the Italian Ministry of Health (grant no. RC IZS VE 02/23 Eziopatogenesi ed epidemiologia di patogeni emergenti nella filiera avicola: persistenza e adattamento di cloni emergenti di Salmonella Infantis e nuove strategie di biocontrollo).
References
- Franco A, Leekitcharoenphon P, Feltrin F, Alba P, Cordaro G, Iurescia M, et al. Emergence of a clonal lineage of multidrug-resistant ESBL-producing Salmonella Infantis transmitted from broilers and broiler meat to humans in Italy between 2011 and 2014. PLoS One. 2015;10:
e0144802 . DOIPubMedGoogle Scholar - European Food Safety Authority; European Centre for Disease Prevention and Control. The European Union One Health 2019 Zoonoses Report. EFSA J. 2021;19:
e06406 .PubMedGoogle Scholar - Montoro-Dasi L, Lorenzo-Rebenaque L, Marco-Fuertes A, Vega S, Marin C. Holistic strategies to control Salmonella Infantis: an emerging challenge in the European broiler sector. Microorganisms. 2023;11:1765. DOIPubMedGoogle Scholar
- Regulation (EC) No 2160/2003 of the European Parliament and of the Council of 17 November 2003 on the control of salmonella and other specified food-borne zoonotic agents. 2003 [cited 2023 Aug 8]. https://eur-lex.europa.eu/legal-content/EN/ALL/?uri=CELEX:32003R2160.
- Commission Regulation (EU) No 200/2010 of 10 March 2010 implementing Regulation (EC) No 2160/2003 of the European Parliament and of the Council as regards a Union target for the reduction of the prevalence of Salmonella serotypes in adult breeding flocks of Gallus gallus. 2010 [cited 2023 Aug 8]. https://eur-lex.europa.eu/legal-content/en/ALL/?uri=CELEX:32010R0200
- Fitzgerald C, Collins M, van Duyne S, Mikoleit M, Brown T, Fields P. Multiplex, bead-based suspension array for molecular determination of common Salmonella serogroups. J Clin Microbiol. 2007;45:3323–34. DOIPubMedGoogle Scholar
- Yang S-M, Baek J, Kim E, Kim HB, Ko S, Kim D, et al. Development of a genoserotyping method for Salmonella Infantis detection on the basis of pangenome analysis. Microorganisms. 2020;9:67. DOIPubMedGoogle Scholar
- Petrin S, Wijnands L, Benincà E, Mughini-Gras L, Delfgou-van Asch EHM, Villa L, et al. Assessing phenotypic virulence of Salmonella enterica across serovars and sources. Front Microbiol. 2023;14:
1184387 . DOIPubMedGoogle Scholar - European Food Safety Authority, Costa G, Di Piazza G, Koevoets P, Iacono G, Liebana E, et al. Guidelines for reporting whole genome sequencing-based typing data through the EFSA One Health WGS System. EFSA supporting publication. 2022;19:EN-7413. DOIGoogle Scholar
- Lalsiamthara J, Kim JH, Lee JH. Engineering of a rough auxotrophic mutant Salmonella Typhimurium for effective delivery. Oncotarget. 2018;9:25441–57. DOIPubMedGoogle Scholar
- Jiao Y, Guo R, Tang P, Kang X, Yin J, Wu K, et al. Signature-tagged mutagenesis screening revealed a novel smooth-to-rough transition determinant of Salmonella enterica serovar Enteritidis. BMC Microbiol. 2017;17:48. DOIPubMedGoogle Scholar
- Park S, Won G, Kim J, Kim HB, Lee JH. Potent O-antigen-deficient (rough) mutants of Salmonella Typhimurium secreting Lawsonia intracellularis antigens enhance immunogenicity and provide single-immunization protection against proliferative enteropathy and salmonellosis in a murine model. Vet Res (Faisalabad). 2018;49:57. DOIPubMedGoogle Scholar
- Hong Y, Reeves PR. Diversity of o-antigen repeat unit structures can account for the substantial sequence variation of wzx translocases. J Bacteriol. 2014;196:1713–22. DOIPubMedGoogle Scholar
- Liu B, Knirel YA, Feng L, Perepelov AV, Senchenkova SN, Reeves PR, et al. Structural diversity in Salmonella O antigens and its genetic basis. FEMS Microbiol Rev. 2014;38:56–89. DOIPubMedGoogle Scholar
- European Food Safety AuthorityEuropean Centre for Disease Prevention and Control. The European Union Summary Report on Antimicrobial Resistance in zoonotic and indicator bacteria from humans, animals and food in 2018/2019. EFSA J. 2021;19:
e06490 . DOIPubMedGoogle Scholar
Figures
Table
Cite This ArticleOriginal Publication Date: March 18, 2024
1These first authors contributed equally to this article.
2These senior authors contributed equally to this article.
Table of Contents – Volume 30, Number 4—April 2024
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
Sara Petrin, SCS1–Microbiology Department, National and WOAH Reference Laboratory for Salmonella, Istituto Zooprofilattico Sperimentale delle Venezie, 31020 Legnaro (PD), Italy
Top