Early Introduction and Community Transmission of SARS-CoV-2 Omicron Variant, New York, New York, USA
Dakai Liu
1, Yexiao Cheng
1, Hangyu Zhou, Lulan Wang, Roberto Hurtado Fiel, Yehudah Gruenstein, Jean Jingzi Luo, Vishnu Singh, Eric Konadu, Keither James, Calvin Lui, Pengcheng Gao, Carl Urban, Nishant Prasad, Sorana Segal-Maurer, Esther Wurzberger, Genhong Cheng

, Aiping Wu

, and William Harry Rodgers

Author affiliations: NewYork-Presbyterian Queens Hospital, Flushing, New York, USA (D. Liu, J.J. Luo, V. Singh, E. Konadu, K. James, C. Lui, P. Gao, C. Urban, N. Prasad, S. Segal-Maurer, W.H. Rodgers);; Chinese Academy of Medical Sciences & Peking Union Medical College, Beijing, China (Y. Cheng, H. Zhou, A. Wu); Suzhou Institute of Systems Medicine, Suzhou, China (Y. Cheng, H. Zhou, A. Wu);; University of California, Los Angeles, California, USA (L. Wang, G. Cheng); Kaiser Permanente Health, North Valley, California, USA (R.H. Fiel);; LabQ Diagnostics, Brooklyn, New York, USA (Y. Gruenstein, E. Wurzberger); Weill Cornell Medical College, New York (J.J. Luo, C. Urban, N. Prasad, S. Segal-Maurer, W.H. Rodgers)
Main Article
Figure 5
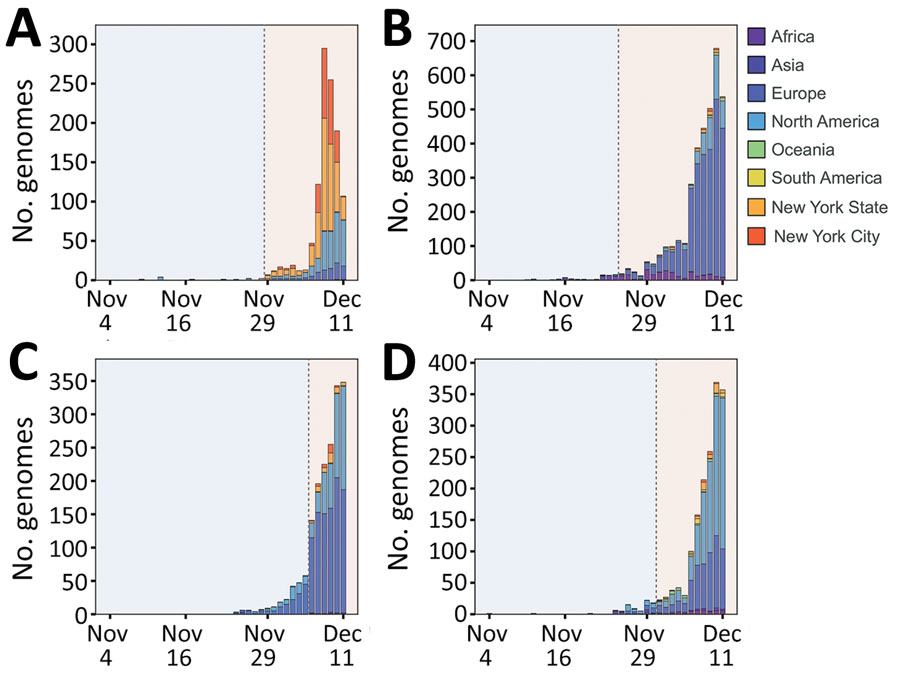
Figure 5. Distribution of SARS-CoV-2 Omicron variant virus isolates clustered into 4 main clades, including viruses identified in this study from New York, New York, USA, November 25–December 11, 2021, and viruses from various regions as obtained from GISAID (https://www.gisaid.org). A) Clade A. B) Clade B. C) Clade C. D) Clade D. For viruses from GISAID, regions were divided into Africa, Asia, Europe, North America (excluding New York state), Oceania, South America, and New York State. Vertical gray dashed lines are to the left of the time at which viruses within the indicated clade were detected in the city of New York during this study. Light blue shading represents the time before our detection of viruses within the indicated clade; light red shading represents the time after we detected the viruses.
Main Article
Page created: January 11, 2023
Page updated: January 21, 2023
Page reviewed: January 21, 2023
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.
