Volume 29, Number 5—May 2023
Dispatch
Novel Circovirus in Blood from Intravenous Drug Users, Yunnan, China
Figure 1
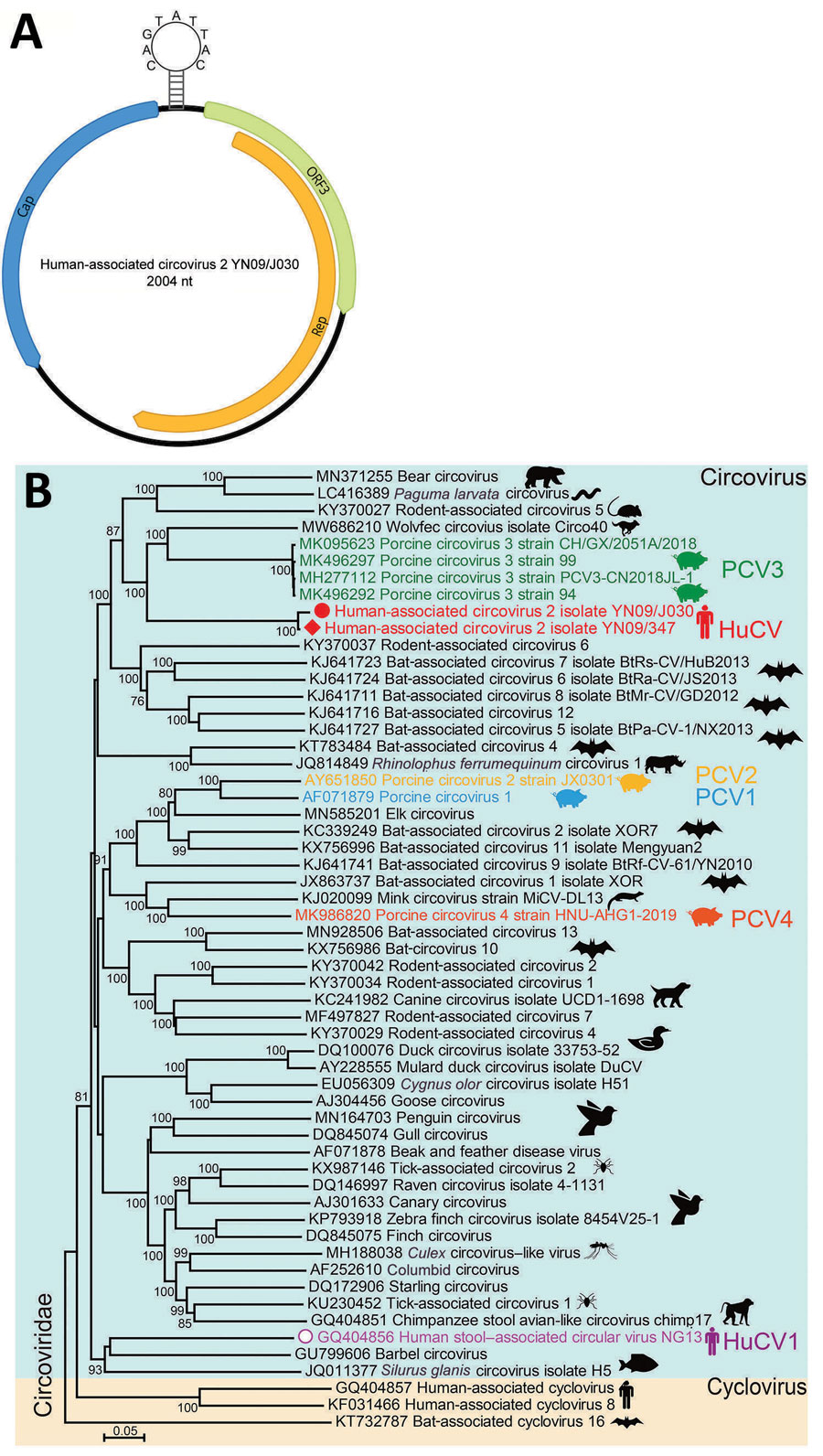
Figure 1. Genomic features of HuCV2 and its phylogenetic relationship with other circoviruses. A) Typical circular genome organization of HuCV2. B) Phylogenetic tree, generated based on the full-length genomic sequences, highlights HuCV2 from 2 patients in China (red circle and red diamond), another circovirus detected in humans (in addition to HuCV1), and the most widely studied known pathogens (PCV1–4). The phylogenetic tree was built in MEGA X (https://www.megasoftware.net) by using the neighbor-joining method. Bootstrap analysis with 1,000 replicates was applied to assess the phylogeny. Scale bar indicates number of nucleotide substitutions per site. Cap, capsid protein; HuCV, human-associated circovirus; ORF3, open reading frame 3; PCV, porcine circovirus; rep, replication-associated protein.