Volume 29, Number 8—August 2023
Dispatch
Candidatus Neoehrlichia mikurensis Infection in Patient with Antecedent Hematologic Neoplasm, Spain1
Figure
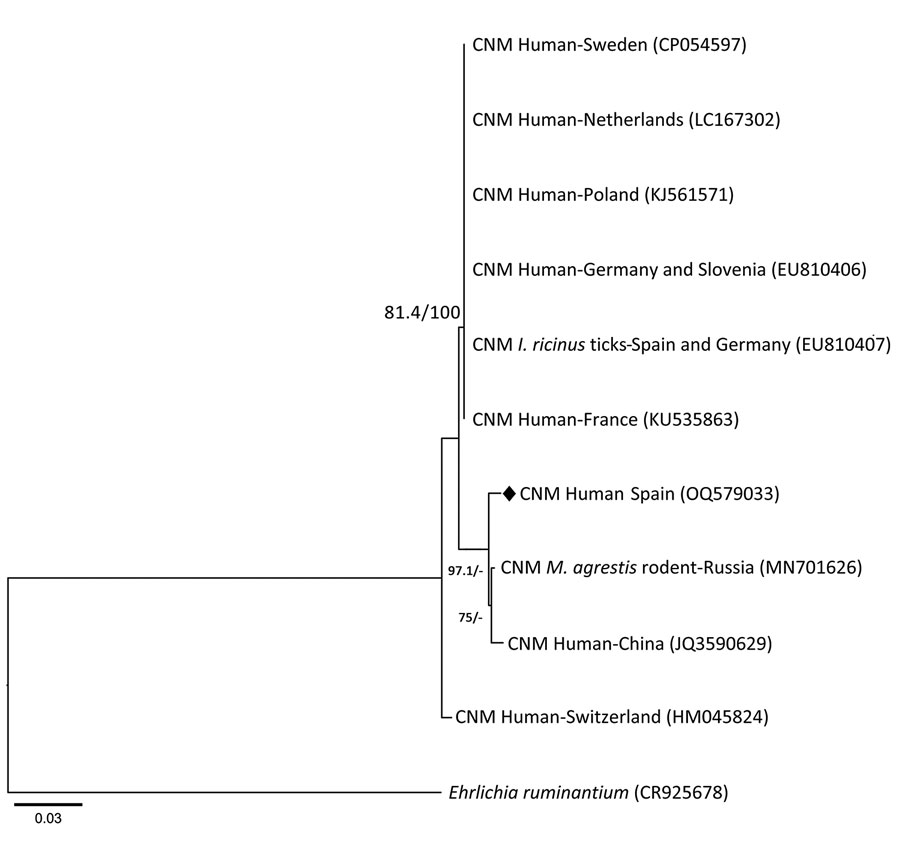
Figure. Phylogenetic analysis of groEL gene from Candidatus Neoehrlichia mikurensis infecting a patient with antecedent hematologic neoplasm, Spain. Phylogenetic tree was generated to compare 809 bp fragments of the 60-kDa heat shock protein gene groEL from Candidatus Neoehrlichia mikurensis by using IQ-tree software version 2.2.0 (http://www.iqtree.org), maximum-likelihood method, and substitution model consisting of 3-parameter model 2 plus empirical base frequencies with rate heterogeneity allowing for a proportion of invariable sites. Values are approximate likelihood ratio test/bootstrap percentages, indicating topologic branch support for maximum-likelihood analysis with 1,000 replicates; values >75% define high stability. Diamond indicates nucleotide sequence of Candidatus N. mikurensis groEL gene fragment obtained in this study. Ehrlichia ruminantium (Anaplasmataceae family) groEL sequence was used as the outgroup. GenBank accession numbers are in parentheses. CNM, Candidatus N. mikurensis; I. ricinus, Ixodes ricinus; M. agrestis, Microtus agrestis. Scale bar indicates nucleotide substitutions per site.
1Data from this study were presented at the joint LXIV National Conference of the Spanish Society of Hematology and Hemotherapy, XXXVIII National Conference of the Spanish Society of Thrombosis and Hemostasis, and 38th World Congress of the International Society of Hematology; October 6–8, 2022; Barcelona, Spain; and International Intracellular Bacteria Meeting; August 23–26, 2022; Lausanne, Switzerland.
2These first authors contributed equally to this article.
3Current affiliation: Hospital Universitario San Agustín, Asturias, Spain.