Volume 29, Number 8—August 2023
Dispatch
Detection of Orientia spp. Bacteria in Field-Collected Free-Living Eutrombicula Chigger Mites, United States
Abstract
Scrub typhus, a rickettsial disease caused by Orientia spp., is transmitted by infected larval trombiculid mites (chiggers). We report the molecular detection of Orientia species in free-living Eutrombicula chiggers collected in an area in North Carolina, USA, to which spotted fever group rickettsiae infections are endemic.
Rickettsioses are distributed worldwide, caused by bacteria in the family Rickettsiaceae, genera Orientia and Rickettsia (1). The pathogens are transmitted by host-feeding arthropods, including ticks, mites, fleas, and lice (2). Among those arthropods are trombiculid mites, which have a widespread global distribution and high species diversity. Among the different stages of the lifecycle of trombiculid mites, only the larvae are ectoparasites, commonly known as chiggers (3). Some species are vectors of intracellular bacterial pathogens in the genus Orientia that causes a potentially lethal human febrile disease, scrub typhus (4,5). Scrub typhus results in considerable illness and death; it causes >1 million cases of illness each year (5). A recent review (6) concluded that trombiculid mites might be widespread vectors of other zoonotic agents not yet recognized. Spotted fever group rickettsiosis diseases, including Rocky Mountain spotted fever, have many of the same symptoms as scrub typhus. A recent review of the literature from 1997–2017 estimated >60% of the rickettsial diseases outside the United States were misdiagnosed (7).
Until recently, scrub typhus was exclusively reported from the so-called tsutsugamushi triangle, stretching from Pakistan in the west to far-eastern Russia in the east to northern Australia in the south. However, scrub typhus was reported recently in the Middle East, southern Chile, and Africa (5,8). The occurrence of scrub typhus pathogens in chiggers in the United States was not investigated. We report the identification of Orientia species in free-living chiggers collected at recreational parks in North Carolina, USA.
In 2022, we collected free-living chiggers using the tile method (9) in different locations in North Carolina: we placed tiles on the ground and then visually inspected for the presence of chiggers after ≈1 minute. When chiggers were present, we collected them with forceps or a small paintbrush and transferred them into vials with 95% ethanol. We identified subsamples from each collection location based on morphological characteristics using published taxonomic keys (10). We identified chiggers as Eutrombicula on the basis of their morphology. In addition, we obtained chigger images of Eutrombicula using scanning electron microscopy at the Analytical Instrumentation Facility at North Carolina State University (Raleigh, North Carolina, USA) (Appendix Figure 1).
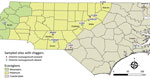
We surface-sterilized individual free-living chiggers and extracted total nucleic acids using the DNeasy Blood & Tissue Kit (QIAGEN, https://www.qiagen.com) (11). In total, 95 chiggers from 10 different locations (10 chiggers/location; 1 location had 5 chiggers) were subjected to microbiome analyses (Figure 1). We randomly selected 8 chiggers for molecular identification using previously described 18S ribosomal RNA gene primers and PCR (12). Amplicons were Sanger sequenced at Eton Bioscience, Inc. (https://www.etonbio.com). The sequences (GenBank accession nos. OQ789321–5) were submitted for BLASTn (https://blast.ncbi.nlm.nih.gov/Blast.cgi) analysis and showed 99–100% identity with homologous sequences of Eutrombicula spp. (accession no. KY922159).
To determine the total bacteria present in the chiggers, we constructed a 16S rRNA sequence library of individual chiggers using the Illumina 16S rRNA (V3-V4 region) metagenomics sequencing library preparation protocol (Illumina, https://www.illumina.com) (13). Sequencing was performed at the University of North Carolina Microbiome Core Facility (Chapel Hill, North Carolina, USA). The Illumina FASTQ files were processed using Quantitative Insights into Microbial Ecology 2 (14) (Appendix). An analysis of amplicon sequence variants (ASVs) revealed that chigger mites contained reads of a bacterial sequence classified as O. tsutsugamushi. O. tsutsugamushi–positive chigger sequence reads were found in 5/10 sampling sites as follows: 3 sites in the Piedmont region (Falls Lake State Recreation Area [1 positive/10 chiggers], Kerr Lake State Recreation Area [8 positive/10 chiggers], and Morrow Mountain State Park [1 positive/10 chiggers]); and 2 sites in the Coastal Plains region (Lumber River State Park [9 positive/10 chiggers] and Merchant Millpond State Park [1 positive/5 chiggers]) (Figure 1). By using the Greengenes 16S rRNA database, we found 13 ASVs to O. tsutsugamushi. To further confirm those results, we extracted representative sequences from the sequencing data and conducted BLASTn searches against the National Center for Biotechnology Information (NCBI) databases. We found 12 ASVs showed a high nucleotide identity (99.5%–100%) to the O. tsutsugamushi strain Kato (D38624), isolated from a human scrub typhus case in Kurosawa village, Japan (https://u.osu.edu/scrubtyphus/the-kato-strain). One ASV exhibited 89.46% homology to an O. tsutsugamushi sequence that was excluded from the phylogenetic analysis. We then examined the approximate phylogenetic relationships for 12 of the O. tsutsugamushi sequence variants from our study sites with other O. tsutsugamushi sequences obtained from the NCBI database using BLASTn (accessed January 18, 2023). We performed multiple alignments with the ClustalW program and maximum-likelihood method tree with the Kimura 2-parameter method from the MEGA X (https://www.megasoftware.net) software package. The phylogenetic analysis revealed that all 12 of the O. tsutsugamushi ASVs clustered closely to O. tsutsugamushi strains from Asia (Figure 2).
To further verify the identity of O. tsutsugamushi detected in our free-living chigger samples, we amplified a 47-kDa htrA (high-temperature requirement A) gene in the 20 chigger samples that were positive for the O. tsutsugamushi 16S rRNA gene. The primers for the first round of PCR were Ot-145F and Ot-1780R, and for the second round, Ot-263F and Ot-1133R (15) (Appendix). The 47-kDa gene amplification products were Sanger sequenced at Eton Bioscience. Four samples (FC28, 36, 38, 109) were 92.6%–97.29% identical to the O. tsutsugamushi HN82 strain (GenBank accession no. LC431268) and 92%–97% identical to the O. tsutsugamushi Kato strain (accession no. LS398550) after trimming low-quality sequences. Sixteen samples yielded ambiguous sequences, suggesting the presence of multiple Orientia species or primer binding sites caused by high variation in the 47 kDa gene among Orientia species in our samples. Jiang et al. (15) studied the genetic variation of Orientia in this region of the genome and reported percent identity of 17 isolates of Orientia as 82.2%–83.3% (15). Among our 4 Orientia 47 kDa sequences from North Carolina chiggers, identity was 93.97%–98%. Phylogenetic analysis revealed that all 4 of those O. tsutsugamushi sequences clustered to O. tsutsugamushi strains from Asia (Appendix Figure 2).
This study identified Orientia species within the United States in free-living Eutrombicula chiggers that were collected in North Carolina. This result is epidemiologically significant because it indicates vertical circulation of Orientia species in chiggers collected within the continental United States. The presence of Orientia species in free-living larvae suggests that the bacteria are maintained through transovarial transmission. Further studies are needed to complete sequencing of the 47-kDa htrA gene (htrA) in our samples, determine how widely distributed Orientia spp.–infected free-living and host-attached chiggers are in the United States, and ascertain whether wild animals that serve as hosts for chiggers become infected and infectious and develop symptoms of illness. Clinicians in this region should be alert for possible human cases of illness resulting from Orientia spp. infection.
Dr. Chen is a postdoctoral research scholar in the Department of Entomology and Plant Pathology at North Carolina State University. Her research interests include vector-borne diseases, microbiomes, and vector control.
Acknowledgment
This research was supported by a grant from the National Institutes of Health, National Institute of Allergy and Infectious Diseases (grant no. 1R03AI166406-01). L.P. was supported by a grant from Southeast Center for Agricultural Health and Injury Prevention. This research was also supported by a grant from the Department of the Army, US Army Contracting Command, Aberdeen Proving Ground, Natick Contracting Division, Fort Detrick, Maryland, under a Deployed Warfighter Protection (DWFP) program grant (no. W911QY1910003 awarded to R.M.R.).
References
- Abdad MY, Abou Abdallah R, Fournier P-E, Stenos J, Vasoo S. A concise review of the epidemiology and diagnostics of rickettsioses: Rickettsia and Orientia spp. J Clin Microbiol. 2018;56:
e01728-17 . DOIPubMedGoogle Scholar - Blanton LS. The rickettsioses: a practical update. Infect Dis Clin North Am. 2019;33:213–29. DOIPubMedGoogle Scholar
- Chen K, Roe RM, Ponnusamy L. Biology, systematics, microbiome, pathogen transmission and control of chiggers (Acari: Trombiculidae, Leeuwenhoekiidae) with emphasis on the United States. Int J Environ Res Public Health. 2022;19:15147. DOIPubMedGoogle Scholar
- Abarca K, Martínez-Valdebenito C, Angulo J, Jiang J, Farris CM, Richards AL, et al. Molecular description of a novel Orientia species causing scrub typhus in Chile. Emerg Infect Dis. 2020;26:2148–56. DOIPubMedGoogle Scholar
- Xu G, Walker DH, Jupiter D, Melby PC, Arcari CM. A review of the global epidemiology of scrub typhus. PLoS Negl Trop Dis. 2017;11:
e0006062 . DOIPubMedGoogle Scholar - Weitzel T, Makepeace BL, Elliott I, Chaisiri K, Richards AL, Newton PN. Marginalized mites: neglected vectors of neglected diseases. PLoS Negl Trop Dis. 2020;14:
e0008297 . DOIPubMedGoogle Scholar - van Eekeren LE, de Vries SG, Wagenaar JFP, Spijker R, Grobusch MP, Goorhuis A. Under-diagnosis of rickettsial disease in clinical practice: a systematic review. Travel Med Infect Dis. 2018;26:7–15. DOIPubMedGoogle Scholar
- Acosta-Jamett G, Martínez-Valdebenito C, Beltrami E, Silva-de La Fuente MC, Jiang J, Richards AL, et al. Identification of trombiculid mites (Acari: Trombiculidae) on rodents from Chiloé Island and molecular evidence of infection with Orientia species. PLoS Negl Trop Dis. 2020;14:
e0007619 . DOIPubMedGoogle Scholar - Upham RW Jr, Hubert AA, Phang OW, Mat YB, Rapmund G. Distribution of Leptotrombidium (Leptotrombidium) arenicola (Acarina: Trombiculidae) on the ground in West Malaysia. J Med Entomol. 1971;8:401–6. DOIPubMedGoogle Scholar
- Wharton GW, Fuller HS. A manual of the chiggers: the biology, classification, distribution and importance to man of the larvae of the family Trombiculidae (Acarina). Mem Entomol Soc Wash. 1952;4:1–185.
- Ponnusamy L, Willcox AC, Roe RM, Davidson SA, Linsuwanon P, Schuster AL, et al. Bacterial microbiome of the chigger mite Leptotrombidium imphalum varies by life stage and infection with the scrub typhus pathogen Orientia tsutsugamushi. PLoS One. 2018;13:
e0208327 . DOIPubMedGoogle Scholar - Otto JC, Wilson K. Assessment of the usefulness of ribosomal 18S and mitochondrial COI sequences in Prostigmata phylogeny. In: Acarology: Proceedings of the 10th International Congress, 2001. Melbourne: CSIRO Publishing Melbourne; 2001.
- Illumina. 16S Metagenomic sequencing library preparation. 2013 [cited 2023 Jun 26]. https://support.illumina.com/downloads/16s_metagenomic_sequencing_library_preparation.html
- Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith GA, et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol. 2019;37:852–7. DOIPubMedGoogle Scholar
- Jiang J, Paris DH, Blacksell SD, Aukkanit N, Newton PN, Phetsouvanh R, et al. Diversity of the 47-kD HtrA nucleic acid and translated amino acid sequences from 17 recent human isolates of Orientia. Vector Borne Zoonotic Dis. 2013;13:367–75. DOIPubMedGoogle Scholar
Figures
Cite This ArticleOriginal Publication Date: July 12, 2023
Table of Contents – Volume 29, Number 8—August 2023
| EID Search Options |
|---|
|
|
|
|
|
|

Please use the form below to submit correspondence to the authors or contact them at the following address:
Loganathan Ponnusamy, Department of Entomology and Plant Pathology, North Carolina State University at Raleigh, 3230 Ligon St, Raleigh, NC 27595, USA
Top