Volume 29, Number 9—September 2023
Dispatch
Rat Hepatitis E Virus in Norway Rats, Ontario, Canada, 2018–2021
Figure 2
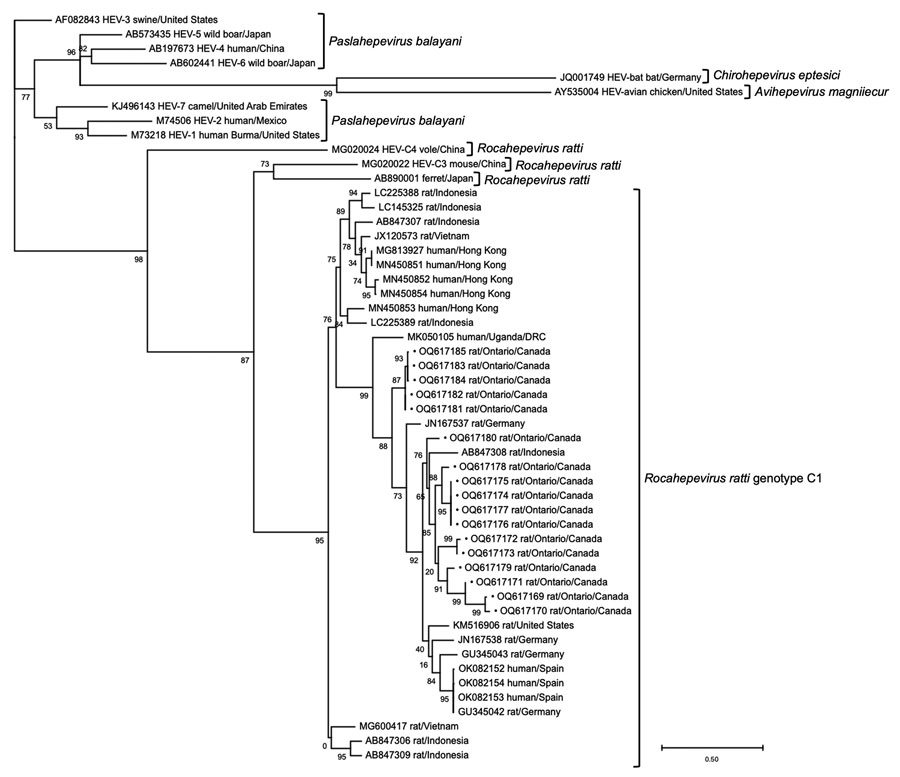
Figure 2. Phylogenetic tree based on the nucleotide alignment of hepatitis E virus (HEV) sequences from rats submitted by pest control professionals in southern Ontario, Canada, during November 2018–June 2021 (black dots), and select reference sequences from other studies in GenBank (accession numbers provided). Maximum-likelihood analysis of a 283-nt fragment of the RNA dependent RNA polymerase of open reading frame 1 was performed by the general time reversible plus gamma plus invariant sites substitution model as determined for the alignment by Smart Model Selection (12). Tree construction was optimized by nearest neighbor interchange and subtree pruning and regrafting with branch support computed by the approximate likelihood-ratio test based on a Shimodaira-Hasegawa-like procedure (13). Only bootstrap values >70% are shown. Scale bar indicates the number of nucleotide substitutions per site. DRC, Democratic Republic of the Congo.
References
- Purdy MA, Drexler JF, Meng XJ, Norder H, Okamoto H, Van der Poel WHM, et al. ICTV virus taxonomy profile: Hepeviridae 2022. J Gen Virol. 2022;103:103. DOIPubMedGoogle Scholar
- Pischke S, Hartl J, Pas SD, Lohse AW, Jacobs BC, Van der Eijk AA. Hepatitis E virus: Infection beyond the liver? J Hepatol. 2017;66:1082–95. DOIPubMedGoogle Scholar
- Meng XJ. From barnyard to food table: the omnipresence of hepatitis E virus and risk for zoonotic infection and food safety. Virus Res. 2011;161:23–30. DOIPubMedGoogle Scholar
- Kenney SP. The current host range of hepatitis E viruses. Viruses. 2019;11:452. DOIPubMedGoogle Scholar
- Wang B, Harms D, Yang XL, Bock CT. Orthohepevirus C: an expanding species of emerging hepatitis E virus variants. Pathogens. 2020;9:154. DOIPubMedGoogle Scholar
- Sridhar S, Yip CCY, Wu S, Chew NFS, Leung KH, Chan JFW, et al. Transmission of rat hepatitis E virus infection to humans in Hong Kong: A clinical and epidemiological analysis. Hepatology. 2021;73:10–22. DOIPubMedGoogle Scholar
- Andonov A, Robbins M, Borlang J, Cao J, Hatchette T, Stueck A, et al. Rat hepatitis E virus linked to severe acute hepatitis in an immunocompetent patient. J Infect Dis. 2019;220:951–5. DOIPubMedGoogle Scholar
- Rivero-Juarez A, Frias M, Perez AB, Pineda JA, Reina G, Fuentes-Lopez A, et al.; HEPAVIR and GEHEP-014 Study Groups. Orthohepevirus C infection as an emerging cause of acute hepatitis in Spain: First report in Europe. J Hepatol. 2022;77:326–31. DOIPubMedGoogle Scholar
- Robinson SJ, Finer R, Himsworth CG, Pearl DL, Rousseau J, Weese JS, et al. Evaluating the utility of pest control sourced rats for zoonotic pathogen surveillance. Zoonoses Public Health. 2022;69:468–74. DOIPubMedGoogle Scholar
- Mulyanto, Suparyatmo JB, Andayani IGAS, Khalid, Takahashi M, Ohnishi H, et al. Marked genomic heterogeneity of rat hepatitis E virus strains in Indonesia demonstrated on a full-length genome analysis. Virus Research. 2014;179:102–12.
- Drexler JF, Seelen A, Corman VM, Fumie Tateno A, Cottontail V, Melim Zerbinati R, et al. Bats worldwide carry hepatitis E virus-related viruses that form a putative novel genus within the family Hepeviridae. J Virol. 2012;86:9134–47. DOIPubMedGoogle Scholar
- Lefort V, Longueville JE, Gascuel O. SMS: smart model selection in PhyML. Mol Biol Evol. 2017;34:2422–4. DOIPubMedGoogle Scholar
- Anisimova M, Gascuel O. Approximate likelihood-ratio test for branches: A fast, accurate, and powerful alternative. Syst Biol. 2006;55:539–52. DOIPubMedGoogle Scholar
- Murphy EG, Williams NJ, Jennings D, Chantrey J, Verin R, Grierson S, et al. First detection of Hepatitis E virus (Orthohepevirus C) in wild brown rats (Rattus norvegicus) from Great Britain. Zoonoses Public Health. 2019;66:686–94. DOIPubMedGoogle Scholar
- Ryll R, Bernstein S, Heuser E, Schlegel M, Dremsek P, Zumpe M, et al. Detection of rat hepatitis E virus in wild Norway rats (Rattus norvegicus) and Black rats (Rattus rattus) from 11 European countries. Vet Microbiol. 2017;208:58–68. DOIPubMedGoogle Scholar