Volume 29, Number 9—September 2023
Dispatch
Anaplasma bovis–Like Infections in Humans, United States, 2015–2017
Figure
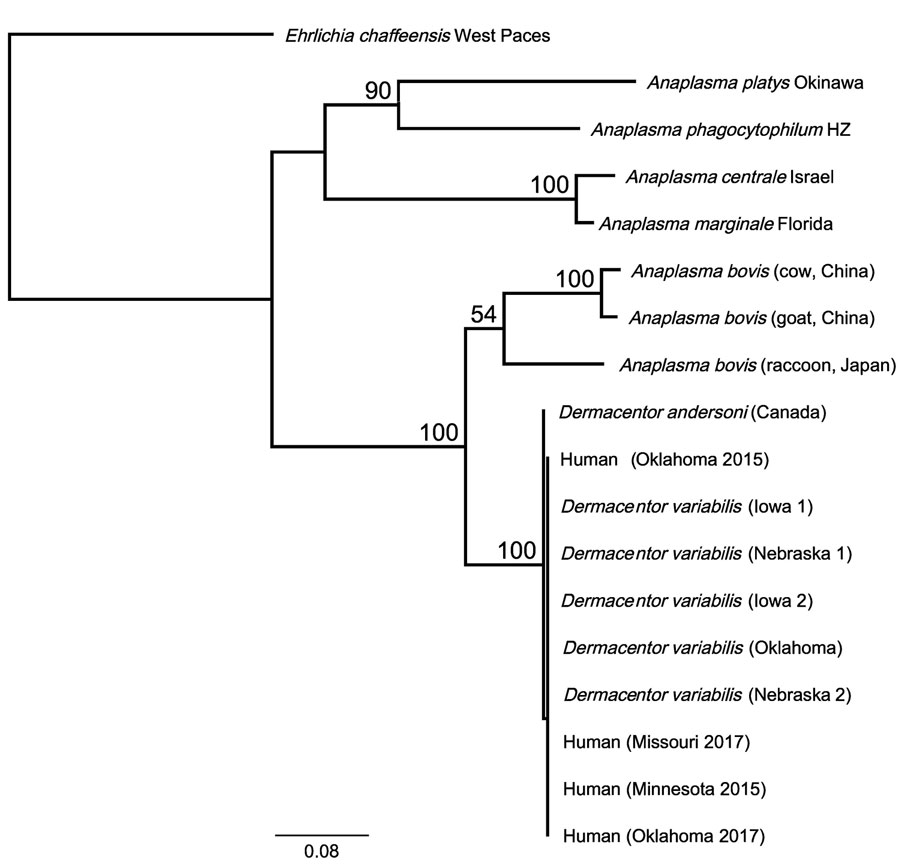
Figure. Phylogenetic relationship of novel human Anaplasma bovis–like pathogen associated with human cases in the United States, 2015–2017, to other A. bovis–like and related Anaplasma species based on 2,039 bp of concatenated rrs, gltA, groEL nucleotide sequences. Phylogenetic relationships were inferred using the RAxML method using the general time reversible plus gamma model (13). One thousand bootstrap replicates were used to estimate the likelihood of the tree; bootstrap values are displayed next to the nodes. Only bootstrap values of >50 are shown. GenBank accession numbers for the samples in this study: OQ772254;, gltA; OQ772255, groEL; and OQ724830, rrs; those for the D. andersoni sample were assigned the following numbers: OQ772256, gltA; OQ772257, groEL; and OQ724821, rrs. Reference sequences from GenBank: Anaplasma bovis (cow, China): MH255937, 16S; MH594290, gltA; MH255906.1, groEL; A. bovis (goat, China): MH255939, 16S; MH255915.1, gltA; MH255907, groEL; A. bovis (raccoon, Japan): GU937020, 16S; JN588561, gltA; JN588562, groEL; Anaplasma platys strain Okinawa: AY077619, 16S; AY077620, gltA; AY077621, groEL; A. phagocytophilum strain HZ NC_007797; A. centrale strain Israel NC_013532; A. marginale strain Florida NC_012026. Ehrlichia chaffeensis strain West Paces (NZ_CP007480) was used as the outgroup. Scale bar represents mean number of nucleotide substitutions per site.
References
- Li H, Zheng Y-C, Ma L, Jia N, Jiang B-G, Jiang R-R, et al. Human infection with a novel tick-borne Anaplasma species in China: a surveillance study. Lancet Infect Dis. 2015;15:663–70. DOIPubMedGoogle Scholar
- Donatien A, Lestoquard F. Rickettsiose bovine Algerienne a R. bovis. Bull Soc Pathol Exot. 1940;33:245–8.
- Lu M, Chen Q, Qin X, Lyu Y, Teng Z, Li K, et al. Anaplasma bovis infection in fever and thrombocytopenia patients—Anhui Province, China 2021. China CDC Wkly. 2022;4:249–53. DOIPubMedGoogle Scholar
- Kingry L, Sheldon S, Oatman S, Pritt B, Anacker M, Bjork J, et al. Targeted metagenomics for clinical detection and discovery of bacterial tick-borne pathogens. J Clin Microbiol. 2020;58:e00147–20. DOIPubMedGoogle Scholar
- Chilton NB, Dergousoff SJ, Lysyk TJ. Prevalence of Anaplasma bovis in Canadian populations of the Rocky Mountain wood tick, Dermacentor andersoni. Ticks Tick Borne Dis. 2018;9:1528–31. DOIPubMedGoogle Scholar
- Lado P, Luan B, Allerdice MEJ, Paddock CD, Karpathy SE, Klompen H. Integrating population genetic structure, microbiome, and pathogens presence data in Dermacentor variabilis. PeerJ. 2020;8:
e9367 . DOIPubMedGoogle Scholar - Lane RS, Mun J, Peribáñez MA, Fedorova N. Differences in prevalence of Borrelia burgdorferi and Anaplasma spp. infection among host-seeking Dermacentor occidentalis, Ixodes pacificus, and Ornithodoros coriaceus ticks in northwestern California. Ticks Tick Borne Dis. 2010;1:159–67. DOIPubMedGoogle Scholar
- Zhuang L, Du J, Cui XM, Li H, Tang F, Zhang PH, et al. Identification of tick-borne pathogen diversity by metagenomic analysis in Haemaphysalis longicornis from Xinyang, China. Infect Dis Poverty. 2018;7:45. DOIPubMedGoogle Scholar
- Sumner JW, Nicholson WL, Massung RF. PCR amplification and comparison of nucleotide sequences from the groESL heat shock operon of Ehrlichia species. J Clin Microbiol. 1997;35:2087–92. DOIPubMedGoogle Scholar
- Rar VA, Livanova NN, Panov VV, Doroschenko EK, Pukhovskaya NM, Vysochina NP, et al. Genetic diversity of Anaplasma and Ehrlichia in the Asian part of Russia. Ticks Tick Borne Dis. 2010;1:57–65. DOIPubMedGoogle Scholar
- Rar V, Livanova N, Tkachev S, Kaverina G, Tikunov A, Sabitova Y, et al. Detection and genetic characterization of a wide range of infectious agents in Ixodes pavlovskyi ticks in Western Siberia, Russia. Parasit Vectors. 2017;10:258. DOIPubMedGoogle Scholar
- Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–3. DOIPubMedGoogle Scholar
- Sashika M, Abe G, Matsumoto K, Inokuma H. Molecular survey of Anaplasma and Ehrlichia infections of feral raccoons (Procyon lotor) in Hokkaido, Japan. Vector Borne Zoonotic Dis. 2011;11:349–54. DOIPubMedGoogle Scholar
- Goethert HK, Telford SR III. Enzootic transmission of Anaplasma bovis in Nantucket cottontail rabbits. J Clin Microbiol. 2003;41:3744–7. DOIPubMedGoogle Scholar
- Yabsley MJ, Romines J, Nettles VF. Detection of Babesia and Anaplasma species in rabbits from Texas and Georgia, USA. Vector Borne Zoonotic Dis. 2006;6:7–13. DOIPubMedGoogle Scholar
1Current affiliation: Agriculture and Agri-food Canada, Lethbridge, Alberta, Canada.
2Current affiliation: Mayo Clinic, Jacksonville, Florida, USA.