Volume 30, Number 2—February 2024
Research
Evidence of Zika Virus Reinfection by Genome Diversity and Antibody Response Analysis, Brazil
Figure 5
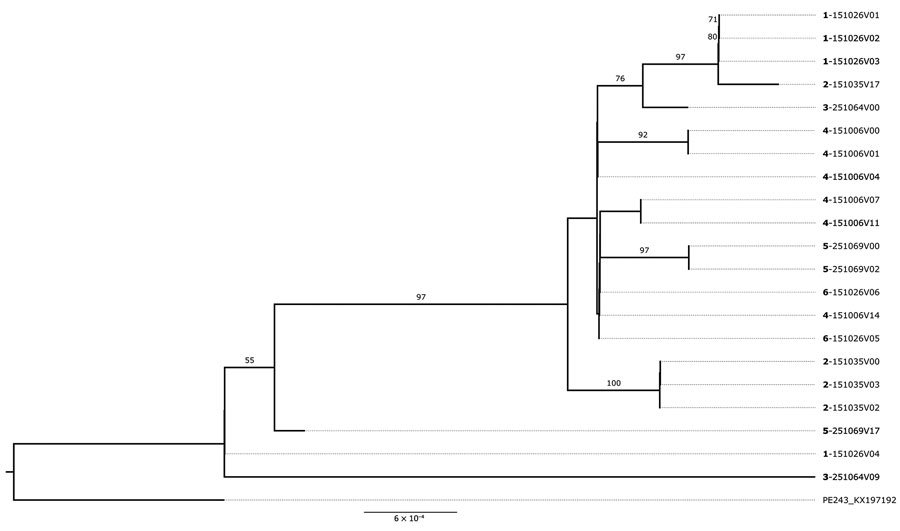
Figure 5. Maximum-likelihood phylogenetic tree supporting Zika virus reinfection among study participants in northern Brazil. The tree shows the 5 participants with divergent samples in which coinfection by different ZIKV genomes was inferred by phylogenetic reconstruction. Divergent samples from the same participant were grouped separately in the tree. Boldface indicates participant identification numbers; visit numbers (V) are indicated. Scale bar indicates number of nucleotide substitutions per site. Numbers on the branches indicate Shimodaira–Hasegawa approximate likelihood ratio test after 1,000 replicates.
1These authors contributed equally to this article.
Page created: January 09, 2024
Page updated: January 24, 2024
Page reviewed: January 24, 2024
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.