Volume 30, Number 3—March 2024
Dispatch
Newly Identified Mycobacterium africanum Lineage 10, Central Africa
Figure
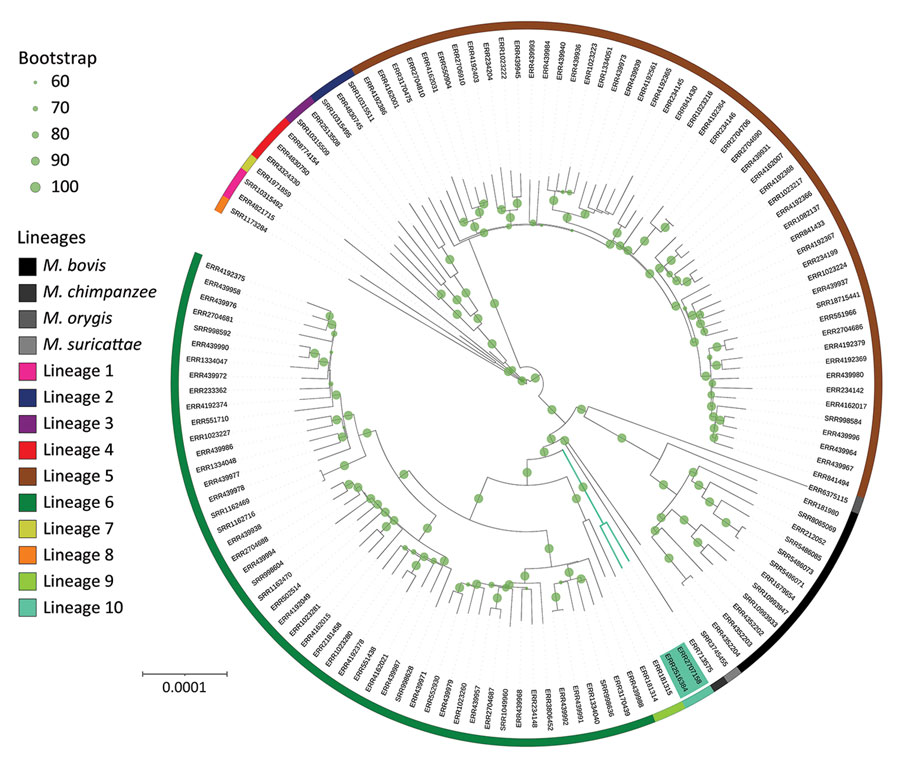
Figure. Global Mycobacterium phylogeny including newly identified M. africanum L10 (proposed) strains (green shading). We selected M. africanum samples for harboring RD9 deletion, having documented country of origin (for the purpose of additional analyses; Appendix 2, Figure 2), and refined our selection to retain a sole representative of each sublineage for each country. This sample represents representing the genetic and geographic diversity of M. africanum in Africa. Specifically for this phylogenetic reconstruction, single-nucleotide polymorphisms were identified in comparison with an M. tuberculosis ancestor (11) and reincorporated into the whole genome to avoid biases in the molecular model or need for Lewis correction. Phylogeny was rooted with M. canettii, subsequently removed for better visualization. Bootstrap support was computed using 100 replicates and shown when ≥0.6. Circles confirm the large support of almost all branches, especially of L10 and its sister branches. L10 branching point lies between L9 and the La_A1 lineage grouping chimpanzee and Dassie bacillus. Scale bar indicates nucleotide substitutions per site.
References
- Gagneux S. Ecology and evolution of Mycobacterium tuberculosis. Nat Rev Microbiol. 2018;16:202–13. DOIPubMedGoogle Scholar
- Ngabonziza JCS, Loiseau C, Marceau M, Jouet A, Menardo F, Tzfadia O, et al. A sister lineage of the Mycobacterium tuberculosis complex discovered in the African Great Lakes region. Nat Commun. 2020;11:2917. DOIPubMedGoogle Scholar
- Coscolla M, Gagneux S, Menardo F, Loiseau C, Ruiz-Rodriguez P, Borrell S, et al. Phylogenomics of Mycobacterium africanum reveals a new lineage and a complex evolutionary history. Microb Genom. 2021;7:00047. DOIPubMedGoogle Scholar
- Napier G, Campino S, Merid Y, Abebe M, Woldeamanuel Y, Aseffa A, et al. Robust barcoding and identification of Mycobacterium tuberculosis lineages for epidemiological and clinical studies. Genome Med. 2020;12:114. DOIPubMedGoogle Scholar
- Phelan JE, O’Sullivan DM, Machado D, Ramos J, Oppong YEA, Campino S, et al. Integrating informatics tools and portable sequencing technology for rapid detection of resistance to anti-tuberculous drugs. Genome Med. 2019;11:41. DOIPubMedGoogle Scholar
- Chen W, Biswas T, Porter VR, Tsodikov OV, Garneau-Tsodikova S. Unusual regioversatility of acetyltransferase Eis, a cause of drug resistance in XDR-TB. Proc Natl Acad Sci U S A. 2011;108:9804–8. DOIPubMedGoogle Scholar
- Dupuy P, Ghosh S, Adefisayo O, Buglino J, Shuman S, Glickman MS. Distinctive roles of translesion polymerases DinB1 and DnaE2 in diversification of the mycobacterial genome through substitution and frameshift mutagenesis. Nat Commun. 2022;13:4493. DOIPubMedGoogle Scholar
- Guyeux C, Sola C, Noûs C, Refrégier G. CRISPRbuilder-TB: “CRISPR-builder for tuberculosis”. Exhaustive reconstruction of the CRISPR locus in mycobacterium tuberculosis complex using SRA. PLOS Comput Biol. 2021;17:
e1008500 . DOIPubMedGoogle Scholar - Couvin D, David A, Zozio T, Rastogi N. Macro-geographical specificities of the prevailing tuberculosis epidemic as seen through SITVIT2, an updated version of the Mycobacterium tuberculosis genotyping database. Infect Genet Evol. 2019;72:31–43. DOIPubMedGoogle Scholar
- Kayomo MK, Mbula VN, Aloni M, André E, Rigouts L, Boutachkourt F, et al. Targeted next-generation sequencing of sputum for diagnosis of drug-resistant TB: results of a national survey in Democratic Republic of the Congo. Sci Rep. 2020;10:10786. DOIPubMedGoogle Scholar
- Comas I, Chakravartti J, Small PM, Galagan J, Niemann S, Kremer K, et al. Human T cell epitopes of Mycobacterium tuberculosis are evolutionarily hyperconserved. Nat Genet. 2010;42:498–503. DOIPubMedGoogle Scholar
- Coscolla M, Lewin A, Metzger S, Maetz-Rennsing K, Calvignac-Spencer S, Nitsche A, et al. Novel Mycobacterium tuberculosis complex isolate from a wild chimpanzee. Emerg Infect Dis. 2013;19:969–76. DOIPubMedGoogle Scholar