Volume 30, Number 3—March 2024
Dispatch
Newly Identified Mycobacterium africanum Lineage 10, Central Africa
Abstract
Analysis of genome sequencing data from >100,000 genomes of Mycobacterium tuberculosis complex using TB-Annotator software revealed a previously unknown lineage, proposed name L10, in central Africa. Phylogenetic reconstruction suggests L10 could represent a missing link in the evolutionary and geographic migration histories of M. africanum.
The traditional view of restricted diversity among bacterial agents causing human and animal tuberculosis is being revised thanks to wide use of whole-genome sequencing (WGS). Besides Mycobacterium canettii, representative of exceptional, nonclonal, early-evolution branching lineages of tubercle bacilli in eastern Africa, several previously unknown lineages of M. tuberculosis complex have been identified in Africa during the past decade. M. tuberculosis complex lineage 7 (L7) was discovered in the Horn of Africa and L8 in the African Great Lakes region (1,2). M. africanum L9 was found only in Djibouti and Somalia. In contrast, 2 other major M. africanum–affiliated lineages contributing substantially to the tuberculosis burden, L5 and L6, are found mostly in western Africa (3). The pathway between eastern and western Africa in the evolutionary history of the bacillus remains unclear. We describe a newly identified sister lineage of L6 and L9 associated with central Africa and discuss implications for determining the evolutionary history of related M. africanum lineages L5, L6, and L9. We based research on publicly available data and thus required no ethics approval.
We used the TB-Annotator platform (G. Senelle, unpub. data, https://www.biorxiv.org/content/10.1101/2023.06.12.526393v1) to integrate WGS data from 102,001 M. tuberculosis complex isolates in the National Center for Biotechnology Information (NCBI) public domain. This platform identifies genetic variations, including single-nucleotide polymorphisms (SNPs), regions of difference (RDs), and IS6110 insertions, differentiating selected genomes from M. tuberculosis H37Rv. The TB-Annotator database also contains information on genotypic drug resistance and geographic location of variant isolation.
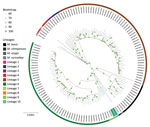
SNPs from an exploratory set comprising 15,699 isolates largely of Africa origin were used to build a phylogenetic tree. Our analysis identified a lineage sister to M. africanum L6 and L9, branching between these lineages and the animal lineage A1 (La_A1) (3). The newly identified lineage is represented by only 2 genomes: ERR2707158, obtained from a strain isolated in 2008 from a patient residing in Kinshasa, Democratic Republic of the Congo (DRC), now incorporated under reference ITM-501386 (CT2008–03226) in the coordinated collections of microorganisms of the Institute of Tropical Medicine (Antwerp, Belgium); and ERR2516384, obtained from a strain isolated in Belgium in 2013 (V. Mathys, pers. comm., email, 2023 Jul 5). The genomes of the new lineage carried none of the SNP markers described in the latest M. tuberculosis complex lineage classification scheme (4) and no SNPs that confer drug resistance.
To confirm the phylogenetic position of those 2 genomes, we identified SNPs from 132 isolates covering the genetic and geographic diversity of L5 and L6 and including representatives of all other lineages using the Genotube pipeline (A. Le Meur, pers. comm., email, 2023 Sep 15) and TB-Profiler (5). Resulting phylogenetic reconstruction confirmed the clustering of ERR2707158 and ERR2516384 in a branch between L6 and L9 and animal lineage La_A1 (Figure). The newly designated L10 samples shared 375 specific SNPs with isolates from our selected set of 132 samples; 243/375 specific SNPs were not detected in any of the 102,001 genomes included in TB-Annotator. Among those specific SNPs, 91 were synonymous (Appendix 1). The pairwise distance between the 2 samples of interest was 382 SNPs (SNPs outside of repetitive regions, manually checked when discordant between 2 pipelines), much shorter than the distance to the other samples of our selection (minimum 1,137 SNPs; average 1,591 ±222 SNPs) (Appendix 2, Figure 1).
We next explored other features of the genomes to corroborate SNP-based phylogenetic inferences. In addition to the deletion of RD9 shared with the L5/L6 branch and animal-associated lineages, the 2 L10 genomes lacked RD7, RD8, and RD10 (3). However, they did not show the RD702 (L6/L9) or RD713 (L5) deletions. In contrast, the 2 unclassified genomes harbored the same specific large 9,134 nt deletion (Rv0613c–Rv0622) in M. tuberculosis H37Rv (NC\_000962.3:706602–715736) not observed in any other lineage. This segment included the toxin/antitoxin gene pair vapB29/vapC29. Two other shared deletions encompassed eis and dnaE2 (Appendix 1), potentially limiting the ability to acquire aminoglycoside resistance (6) and possibly affecting some mutational properties (7) of those M. africanum strains. The 2 genomes also shared 4 IS6110 copies at a position found in no other lineage (Appendix 1). In the CRISPR locus of the 2 L10 genomes, reconstructed using CRISPRbuilder-TB (8), we found the same absence of spacers 7 and 9 (43-spacer spoligotype format) seen in L6, L9, and La_A1 (Table) and all last spacers starting from spacers 22 (ERR2516384) or 26 (ERR2707158) (Table).
The genetic features of the strains we identified, combining outlying phylogenetic position, genetic distance from the L6/L9 branch and other known M. tuberculosis lineages, distinctive regions of deletions and IS6110 insertions, and specific spoligotype signatures, led us to propose their classification in a newly designated L10 lineage. We propose 3 synonymous SNPs (gyrA G7901T, recN C1920096T, and dnaG C2621730T) compared with the H37Rv 000962.3 reference sequence in housekeeping genes to identify the new lineage.
To evaluate potential regional and global circulation of L10 strains, we searched for similar spoligotype patterns using SITVIT2, which accumulates spoligotypes from >110,000 isolates from 131 countries (9). We identified a single instance, BEL04200301729, showing the same spoligotype pattern as ERR2516384, which might represent a third occurrence of L10. Of note, that strain was isolated in the Republic of the Congo, a country neighboring DRC, where ERR2707158 was collected (Appendix 2, Figure 2). We also browsed spoligotyping results from next generation sequencing data, collected from ≈1,500 isolates from a 2016–2017 national survey in DRC, targeted using Deeplex Myc-TB (https://www.deeplex.com) (10) but detected no similar pattern. Thus, both global (TB-Annotator and SITVIT2) and local (10) datasets suggested that L10 strains are rare at the worldwide level, and aside from migratory dissemination, likely restricted to central Africa. Mapping of M. africanum diversity in Africa showed that in addition to L10, central Africa also hosts a relatively large diversity of L5 strains (Appendix 2, Figure 2).
Despite the rarity of L10, its specific phylogenetic positioning and presence in central Africa provide new elements to the complex evolutionary history of M. africanum. Currently, the most likely scenario favors western Africa as the place of origin of all M. africanum variants (3). This scenario implies that L5 and L6 ancestors emigrated from eastern Africa and diversified in western Africa and that L9 migrated back to eastern Africa. Finding L10 in central Africa with intermediate branching between L5 and L6/L9 can fit this scenario but adds an independent migration from western Africa to central Africa. Alternatively, M. africanum could have emerged close to central Africa and subsequently migrated westwards and eastwards. This alternative scenario, however, would require greater sampling in central regions of Africa to gain real support.
Through the extensive mining of WGS and genotyping databases, we newly identified a thus far rare M. tuberculosis complex lineage, L10 (proposed), present in central Africa. The lineage is characterized by a new region of deletion, IS6110 insertions, and 243 SNPs, including gyrA G7901T, recN C1920096T, and dnaG C2621730T. L10 represents a sister clade to L6, found mainly in western Africa, and L9, specifically in eastern Africa, and reveals a putative previously missing piece in the evolutionary history and migrations of M. africanum. Our findings extend the known diversity of M. africanum in Africa.
Dr. Guyeux is a professor of computer science at the Franche-Comté Électronique Mécanique Thermique et Optique—Sciences et Technologies Institute, University of Franche-Comté in Belfort, France. His research interests include microbial evolution, with a particular focus on extensive sets of genomes.
Acknowledgment
We thank the Institute of Tropical Medicine of Antwerp (Belgium) and Genoscreen (France) for sharing their SRA data on public databases. We thank Vanessa Mathys for providing location of isolation for ERR2516384 sample.
References
- Gagneux S. Ecology and evolution of Mycobacterium tuberculosis. Nat Rev Microbiol. 2018;16:202–13. DOIPubMedGoogle Scholar
- Ngabonziza JCS, Loiseau C, Marceau M, Jouet A, Menardo F, Tzfadia O, et al. A sister lineage of the Mycobacterium tuberculosis complex discovered in the African Great Lakes region. Nat Commun. 2020;11:2917. DOIPubMedGoogle Scholar
- Coscolla M, Gagneux S, Menardo F, Loiseau C, Ruiz-Rodriguez P, Borrell S, et al. Phylogenomics of Mycobacterium africanum reveals a new lineage and a complex evolutionary history. Microb Genom. 2021;7:00047. DOIPubMedGoogle Scholar
- Napier G, Campino S, Merid Y, Abebe M, Woldeamanuel Y, Aseffa A, et al. Robust barcoding and identification of Mycobacterium tuberculosis lineages for epidemiological and clinical studies. Genome Med. 2020;12:114. DOIPubMedGoogle Scholar
- Phelan JE, O’Sullivan DM, Machado D, Ramos J, Oppong YEA, Campino S, et al. Integrating informatics tools and portable sequencing technology for rapid detection of resistance to anti-tuberculous drugs. Genome Med. 2019;11:41. DOIPubMedGoogle Scholar
- Chen W, Biswas T, Porter VR, Tsodikov OV, Garneau-Tsodikova S. Unusual regioversatility of acetyltransferase Eis, a cause of drug resistance in XDR-TB. Proc Natl Acad Sci U S A. 2011;108:9804–8. DOIPubMedGoogle Scholar
- Dupuy P, Ghosh S, Adefisayo O, Buglino J, Shuman S, Glickman MS. Distinctive roles of translesion polymerases DinB1 and DnaE2 in diversification of the mycobacterial genome through substitution and frameshift mutagenesis. Nat Commun. 2022;13:4493. DOIPubMedGoogle Scholar
- Guyeux C, Sola C, Noûs C, Refrégier G. CRISPRbuilder-TB: “CRISPR-builder for tuberculosis”. Exhaustive reconstruction of the CRISPR locus in mycobacterium tuberculosis complex using SRA. PLOS Comput Biol. 2021;17:
e1008500 . DOIPubMedGoogle Scholar - Couvin D, David A, Zozio T, Rastogi N. Macro-geographical specificities of the prevailing tuberculosis epidemic as seen through SITVIT2, an updated version of the Mycobacterium tuberculosis genotyping database. Infect Genet Evol. 2019;72:31–43. DOIPubMedGoogle Scholar
- Kayomo MK, Mbula VN, Aloni M, André E, Rigouts L, Boutachkourt F, et al. Targeted next-generation sequencing of sputum for diagnosis of drug-resistant TB: results of a national survey in Democratic Republic of the Congo. Sci Rep. 2020;10:10786. DOIPubMedGoogle Scholar
- Comas I, Chakravartti J, Small PM, Galagan J, Niemann S, Kremer K, et al. Human T cell epitopes of Mycobacterium tuberculosis are evolutionarily hyperconserved. Nat Genet. 2010;42:498–503. DOIPubMedGoogle Scholar
- Coscolla M, Lewin A, Metzger S, Maetz-Rennsing K, Calvignac-Spencer S, Nitsche A, et al. Novel Mycobacterium tuberculosis complex isolate from a wild chimpanzee. Emerg Infect Dis. 2013;19:969–76. DOIPubMedGoogle Scholar
Figure
Table
Cite This ArticleOriginal Publication Date: February 01, 2024
Table of Contents – Volume 30, Number 3—March 2024
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
Guislaine Refrégier, Ecologie Systématique et Evolution, 12 rue 128, 91190, Gif-sur-Yvette, France
Top