Volume 30, Number 4—April 2024
Research
A One Health Perspective on Salmonella enterica Serovar Infantis, an Emerging Human Multidrug-Resistant Pathogen
Figure 4
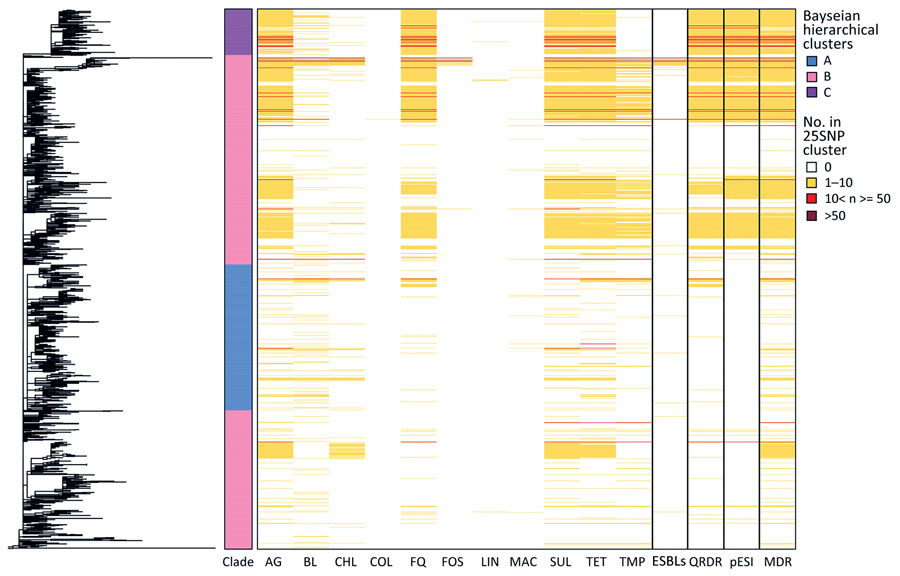
Figure 4. Phylogeny and heatmap of antimicrobial resistance and pESI in study of One Health perspective of emerging multidrug-resistant pathogen Salmonella enterica serovar Infantis. Heatmap shows the number of isolates in each 25 single-nucleotide polymorphism representative cluster (n = 1,288) of the eBG31 maximum-likelihood phylogeny with genes conferring resistance to antimicrobial drugs. Fastbaps clade and the number of isolates with MDR, ESBLs, mutations in the QRDR conferring resistance to fluoroquinolones and pESI presence are also shown. AG, aminoglycosides; BL, β-lactams; CHL, chloramphenicol; COL, colistin; ESBLs, extended β-lactamases; FQ, fluoroquinolones; FOS, fosfomycin; LIN, lincosamides; MAC, macrolides; MDR, multidrug-resistant; SUL, sulphonamides; TET, tetracyclines; TMP, trimethoprim.
1Current affiliation: University of Edinburgh, Edinburgh, Scotland, UK.
2Current affiliation: Utrecht University, Utrecht, the Netherlands.
3Current affiliation: UK Health Security Agency, London, UK.
4Current affiliation: European Bioinformatics Institute, Cambridge, UK.