Volume 21, Number 8—August 2015
Dispatch
Detection and Full-Length Genome Characterization of Novel Canine Vesiviruses
Figure 2
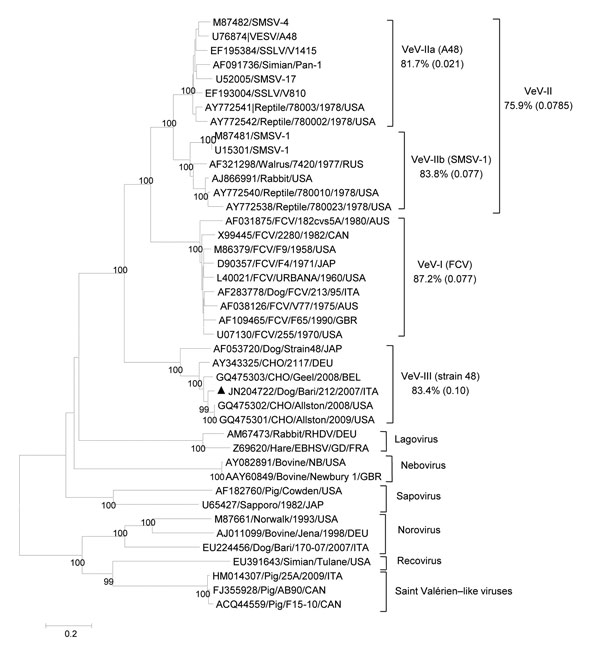
Figure 2. Phylogenetic tree based on the complete amino acid sequence of the capsid protein of vesiviruses (VeVs). The tree was constructed by using a selection of feline calicivirus strains and all of the VeV strains available in the GenBank database. In addition, viruses representative of the other established and candidate calicivirus genera were included. VeV groups were defined on the basis of distance matrix comparison and phylogenetic clustering. The mean identity among strains of the main genetic groups (indicated by Roman numerals and a letter or both) is shown. Numbers in parentheses indicate SDs. Black triangle indicates the canine VeV strain Bari/212/07/ITA. DEU, Germany; EBHSV, European brown hare syndrome virus; SMSV, San Miguel sea lion virus; SSLV, stellar sea lion virus; RHDV, rabbit hemorrhagic disease virus; VESV, vesicular exanthema of swine virus. Scale bar represents the number of amino acid substitutions per site.