Genomic Microevolution of Vibrio cholerae O1, Lake Tanganyika Basin, Africa
Yaovi M.G. Hounmanou, Elisabeth Njamkepo, Jean Rauzier, Karin Gallandat, Aurélie Jeandron, Guyguy Kamwiziku, Klaudia Porten, Francisco Luquero, Aaron Aruna Abedi, Baron Bashige Rumedeka, Berthe Miwanda, Martin Michael, Placide Welo Okitayemba, Jaime Mufitini Saidi, Renaud Piarroux, François-Xavier Weill, Anders Dalsgaard, and Marie-Laure Quilici

Author affiliations: University of Copenhagen, Frederiksberg, Denmark (Y.M.G. Hounmanou, A. Dalsgaard); Institut Pasteur, Université Paris Cité, Paris, France (E. Njamkepo, J. Rauzier, F.-X. Weill, M.-L. Quilici); London School of Hygiene and Tropical Medicine, London, UK (K. Gallandat, A. Jeandron); University of Kinshasa, Kinshasa, Democratic Republic of the Congo (G. Kamwiziku); Epicentre, Paris (K. Porten, F. Luquero); Ministry of Public Health, Kinshasa (A. Aruna Abedi, P. Welo Okitayemba); Ministry of Public Health, Uvira, Democratic Republic of the Congo (B. Bashige Rumedeka, J. Mufitini Saidi); Institut National de Recherche Biomédicale, Kinshasa (B. Miwanda); Sokoine University of Agriculture College of Veterinary Medicine and Biomedical Sciences, Morogoro, Tanzania (M. Michael); Sorbonne Université, Inserm UMR 1136, Assistance Publique-Hôpitaux de Paris, Hôpital Pitié-Salpêtrière, Paris (R. Piarroux)
Main Article
Figure 1
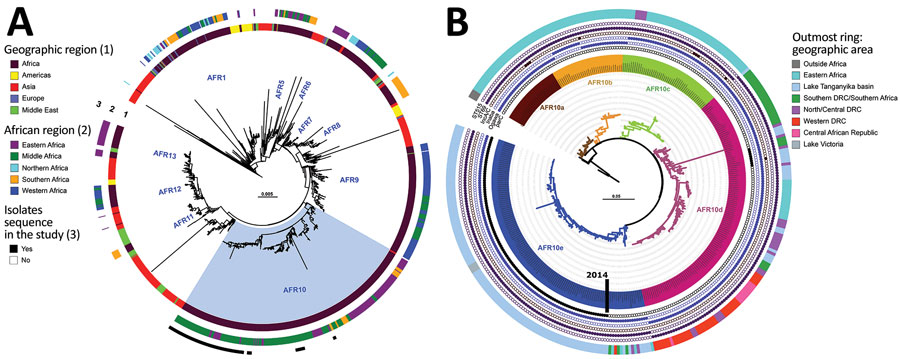
Figure 1. Phylogenomics of clinical and environmental Vibrio cholerae O1 El Tor isolates from the Lake Tanganyika basin, Africa. A) Maximum-likelihood phylogeny of 1,366 seventh pandemic V. cholerae O1 El Tor (7PET) genomes with strain A6 as the outgroup. The different sublineages introduced into Africa are indicated. Light blue indicates AFR10 sublineage. Rings 1 and 2 show geographic origin of isolates; ring 3 shows isolates sequenced in this study. B) Maximum-likelihood tree for 357 AFR10 isolates, with strain N16961 as an outgroup. The 5 clades are color-coded: AFR10a, brown; AFR10b, yellow; AFR10c, green; AFR10d, pink; and AFR10e, blue. The outermost ring indicates the geographic locations of the different isolates in the tree. Filled circles indicate the presence of ST69 or ST515, Ogawa and Inaba serotypes, IncA/C plasmid, and the S85L mutation in parC; open circles indicate their absence. MLST, multilocus sequence typing; ST, sequence type.
Main Article
Page created: December 09, 2022
Page updated: December 22, 2022
Page reviewed: December 22, 2022
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.
