Volume 29, Number 1—January 2023
Dispatch
Plasmodium falciparum pfhrp2 and pfhrp3 Gene Deletions in Malaria-Hyperendemic Region, South Sudan
Abstract
Pfhrp2 and pfhrp3 gene deletions threaten the use of Plasmodium falciparum malaria rapid diagnostic tests globally. In South Sudan, deletion frequencies were 15.6% for pfhrp2, 20.0% for pfhrp3, and 7.5% for double deletions. Deletions were approximately twice as prevalent in monoclonal infections than in polyclonal infections.
Histidine-rich protein 2 (HRP2) is the primary target of the Plasmodium falciparum rapid diagnostic tests (RDT) that are a cornerstone of malaria control efforts in the high-burden, low-resource contexts in which malaria mortality is most acute (1). Increasing prevalence of P. falciparum parasites that do not express HRP2 or its paralogue histidine-rich protein 3 (encoded by the pfhrp2 and pfhrp3 genes) are affecting the accuracy of the RDTs. Infections with pfhrp2 deletions are missed by HPR2-based RDTs much of the time; infections with double deletions (missing both pfhrp2 and pfhrp3 genes) are invisible to RDTs and create false-negative results. Because these deletions represent an existential threat to recent gains made in malaria control, the World Health Organization (WHO) has emphasized the critical need for surveillance (2).
Malaria is a leading cause of illness and death in South Sudan (3), where insufficient malaria prevention activities and a lack of access to healthcare combine dangerously. Despite the geographic and strategic importance of South Sudan in East and Central Africa, the only evidence of pfhrp2 and pfhrp3 deletions from the country come from a single report confirming their presence in 3 travelers to Australia (4). Accurate estimates of deletions could help responders delineate factors associated with deletions, predict future RDT needs, and clarify dynamics of false negativity rates in South Sudan overall.
In 2019, in collaboration with the South Sudanese Ministry of Health, Médecins Sans Frontières began a seasonal malaria chemoprophylaxis campaign combined with an assessment of molecular markers of antimalaria drug resistance in Yambio County, a malaria-endemic region of Western Equatoria State. We describe the frequency of pfhrp2 and pfhrp3 and double deletions in this clinical cohort, as well as the association between deletions, demographic factors, and infection characteristics in South Sudan. This study was approved by the internal ethics review board at Médecins Sans Frontières and by the South Sudan Research Ethics Committee. All participants provided informed consent.
We analyzed finger-prick blood samples collected in 9 villages in Yambio at the end of the malaria peak (January–February 2020) from persons >6 months of age with symptomatic malaria infection positively diagnosed by pan-pLDH–based RDT (CareStart Malaria PAN [pLDH] Ag RDT; Access Bio, https://accessbio.net). We performed malaria confirmation and speciation of 594 dried blood spot samples by multiplex PCR (5). Confirmed P. falciparum samples with high DNA quality (n = 518) underwent genotyping and molecular analysis for deletions in exon 2 of pfhrp2 and pfhrp3 (5) (Appendix). Demographic information was collected for all samples (Appendix). We defined multiplicity of infection (MOI) as the number of parasitic genotypes per infection and analyzed for a random subsample (n = 419) by amplifying P. falciparum merozoite surface protein 1 and 2 genes (pfmsp1, pfmsp2) (6). We defined monoclonal infection as the detection of a single PCR fragment for each locus and polyclonal infection as the detection of >1 PCR fragment for >1 locus.
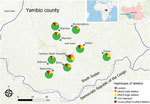
Overall deletion frequency (including samples with both single and double deletions) among 518 genotyped PCR-positive samples was 15.6% for pfhrp2 and 20.0% for pfhrp3 (Table 1). Double deletions were found in 7.5% of isolates; patients at Yambio State Hospital had nearly twice the rate of double deletions (15.9%) as patients at all other sites (Figure 1). In 7/9 study sites, >10% of samples did not amplify pfhrp2; >5% of isolates were double-deleted in nearly half (4/10) of sites. Pfhrp2 deletion rates in South Sudan were as high as or higher than the country’s immediate neighbors, where reported deletion rates from specific sites have varied from 26% in Ethiopia to 19% in Kenya, 6% in the Democratic Republic of the Congo, 3% in Uganda, and <1% in Sudan (7–11).
Monoclonality was the only factor significantly associated with both pfhrp2 and pfhrp3 deletions and double deletions (Table 2). Even so, the frequency of deletion among polyclonal infections was higher than expected. Severe malaria cases exhibited significantly more pfhrp3 and double deletions than uncomplicated infections. Patients >14 years of age were more likely to harbor deletions than were patients <5 and 5–14 years of age, although the difference was significant only for pfhrp3 (Table 2).
Unequal deletion distribution between proximate geographic zones has been previously reported (9,10), and variability in double deletion rates by study site in Yambio was notable, suggesting possible hot spots. However, the higher rates seen at Yambio State Hospital could also be explained by its significantly higher proportion of monoclonal infections (Appendix). The small sample sizes at individual sites leave conclusions about hot spots unsettled, but they underscore the importance of realistic malaria control strategies targeted to the local molecular marker landscape.
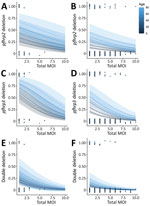
We used a statistically significant multivariable logistic regression model for total MOI, infection severity, and patient age factors for pfhrp2, pfhrp3 and double deletions (Figure 2). Age was the only significant predictor of pfhrp2 deletions (after adjusting for MOI and severity of infection), whereas age, MOI, and clinical severity were all predictors for pfhrp3 deletions. Only MOI was a significant predictor of double deletions (Appendix).
Most research surrounding the interaction between MOI and pfhrp2 and pfhrp3 deletions concludes that polyclonal infections mask deletions and lead to underestimates in deletion prevalence (12). Our results support this conclusion, finding lower MOI in Yambio associated with most deletions. In high-transmission settings, younger persons tend to have higher MOIs (13). We also found lower MOI and older patient age associated with deletions, contrasting with studies that have linked deletions to lower age but failed to consider MOI as a confounder (9).
In addition, the fitness-cost of deletions (the effects on the parasite after losing 1 of its proteins) could be another way that age, disease severity, and deletion risk interact, because milder disease has previously been associated with pfhrp2 deletion (14) and persons acquire immunity against P. falciparum as they age (13). In this area, our cohort breaks with consensus, finding deletions more commonly in complicated malaria patients. We believe this difference might reflect the difficulties of diagnosing febrile disease in South Sudan, where the signs of severe malaria might be caused by other undetected infections.
Sample collection in this study occurred at the end of the high-intensity malaria transmission season, when potentially high parasitic diversity but low prevalence could favor spread of gene-deleted organisms, making deletions easier to detect (14). When interpreting pfhrp2 deletion surveillance, the transmission period should be considered (9,15).
This study was limited because it was a secondary analysis of a study of molecular markers of antimalarial drug resistance and did not follow WHO protocols for pfhrp2 and pfhrp3 deletion surveillance. Consequently, the precise prevalence of pfhrp2 and pfhrp3 gene deletions causing false-negative results on RDTs in South Sudan was not generated to assess whether it is within the 5% threshold established by WHO (2). We also exclusively used pan p-LDH RDT–positive samples, preventing us from evaluating the effects of deletions on malaria diagnoses.
Characterizing pfhrp2 and pfhrp3 deletions is critical to designing effective public health strategies for malaria control. This study describes these deletions in a clinical cohort in a country with little previous endemic evidence of pfhrp2 and pfhrp3 deletion. Monoclonal infections were a principal predictor of deletions. We identified high levels of single and double deletions of pfhrp2 and pfhrp3, which if more widely present in this or other regions of South Sudan, could s eriously jeopardize HRP2-based RDT effectiveness moving forward. Future studies should be designed according to WHO protocol to produce precise estimates to measure the risk those deletions pose in South Sudan. Local variation in prevalence suggests the potential for deletion hotspots within the country and should be considered when designing malaria control strategies.
Prof. Molina-de la Fuente is a PhD candidate at the University of Alcalá with Dr. Pedro Berzosa as PhD codirector. She is conducting research at the National Centre of Tropical Medicine at Institute of Health Carlos III in Spain on molecular and genomic epidemiology, malaria diagnosis evasion, and antimalaria drug resistance. Dr. Sagrado is the epidemiology adviser in Médecins Sans Frontières OCBA, where she conducts operational and clinical research in infectious and chronic diseases. She also has worked as an epidemiologist at the World Health Organization and the Spanish National Center of Epidemiology’s Institute of Health Carlos III.
Acknowledgments
We thank the population in Yambio region for their participation. We also thank all the Médecins Sans Frontières, Ministry of Health, and partner staff involved in this project. We thank Luz Garcia, Vincenta González, and Rosario Gálvez for laboratory work with samples.
These data were previously presented in poster form at the Spanish Society of Tropical Medicine and International Health Conference in March 2022.
References
- Rogier E, McCaffery JN, Nace D, Svigel SS, Assefa A, Hwang J, et al. Plasmodium falciparum pfhrp2 and pfhrp3 gene deletions from persons with symptomatic malaria infection in Ethiopia, Kenya, Madagascar, and Rwanda. Emerg Infect Dis. 2022;28:608–16. DOIPubMedGoogle Scholar
- World Health Organization. Master protocol for surveillance of pfhrp2/3 deletions and biobanking to support future research [cited 2022 Dec 13]. https://apps.who.int/iris/bitstream/handle/10665/331197/9789240002050-eng.pdf
- Republic of South Sudan Ministry of Health. Malaria Strategic Plan 2021 – 2025. 2020.
- Prosser C, Gresty K, Ellis J, Meyer W, Anderson K, Lee R, et al. Plasmodium falciparum histidine-rich protein 2 and 3 gene deletions in strains from Nigeria, Sudan, and South Sudan. Emerg Infect Dis. 2021;27:471–9. DOIPubMedGoogle Scholar
- Berzosa P, González V, Taravillo L, Mayor A, Romay-Barja M, García L, et al. First evidence of the deletion in the pfhrp2 and pfhrp3 genes in Plasmodium falciparum from Equatorial Guinea. Malar J. 2020;19:99. DOIPubMedGoogle Scholar
- Sondo P, Derra K, Rouamba T, Nakanabo Diallo S, Taconet P, Kazienga A, et al. Determinants of Plasmodium falciparum multiplicity of infection and genetic diversity in Burkina Faso. Parasit Vectors. 2020;13:427. DOIPubMedGoogle Scholar
- Feleke SM, Reichert EN, Mohammed H, Brhane BG, Mekete K, Mamo H, et al. Plasmodium falciparum is evolving to escape malaria rapid diagnostic tests in Ethiopia. Nat Microbiol. 2021;6:1289–99. DOIPubMedGoogle Scholar
- Beshir KB, Sepúlveda N, Bharmal J, Robinson A, Mwanguzi J, Busula AO, et al. Plasmodium falciparum parasites with histidine-rich protein 2 (pfhrp2) and pfhrp3 gene deletions in two endemic regions of Kenya. Sci Rep. 2017;7:14718. DOIPubMedGoogle Scholar
- Parr JB, Verity R, Doctor SM, Janko M, Carey-Ewend K, Turman BJ, et al. Pfhrp2-deleted Plasmodium falciparum parasites in the Democratic Republic of the Congo: a national cross-sectional survey. J Infect Dis. 2017;216:36–44.PubMedGoogle Scholar
- Agaba BB, Anderson K, Gresty K, Prosser C, Smith D, Nankabirwa JI, et al. Molecular surveillance reveals the presence of pfhrp2 and pfhrp3 gene deletions in Plasmodium falciparum parasite populations in Uganda, 2017-2019. Malar J. 2020;19:300. DOIPubMedGoogle Scholar
- Boush MA, Djibrine MA, Mussa A, Talib M, Maki A, Mohammed A, et al. Plasmodium falciparum isolate with histidine-rich protein 2 gene deletion from Nyala City, Western Sudan. Sci Rep. 2020;10:12822. DOIPubMedGoogle Scholar
- Grignard L, Nolder D, Sepúlveda N, Berhane A, Mihreteab S, Kaaya R, et al. A novel multiplex qPCR assay for detection of Plasmodium falciparum with histidine-rich protein 2 and 3 (pfhrp2 and pfhrp3) deletions in polyclonal infections. EBioMedicine. 2020;55:
102757 . DOIPubMedGoogle Scholar - Pinkevych M, Petravic J, Bereczky S, Rooth I, Färnert A, Davenport MP. Understanding the relationship between Plasmodium falciparum growth rate and multiplicity of infection. J Infect Dis. 2015;211:1121–7. DOIPubMedGoogle Scholar
- Watson OJ, Slater HC, Verity R, Parr JB, Mwandagalirwa MK, Tshefu A, et al. Modelling the drivers of the spread of Plasmodium falciparum hrp2 gene deletions in sub-Saharan Africa. eLife. 2017;6:
e25008 . DOIPubMedGoogle Scholar - Koita OA, Ndiaye J, Nwakanma D, Sangare L, Ndiaye D, Joof F. Seasonal changes in the frequency of false negative rapid diagnostic tests based on histidine rich protein 2 (HRP2). Am J Trop Med Hyg. 2013;80:1.
Figures
Tables
Cite This ArticleOriginal Publication Date: December 16, 2022
1These authors contributed equally to this article.
Table of Contents – Volume 29, Number 1—January 2023
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
Maria Jose Sagrado Benito, Médecins Sans Frontières–Barcelona, Carrer de Zamora, 54, 08005 Barcelona, Spain
Top