Volume 29, Number 1—January 2023
Dispatch
Detection of Clade 2.3.4.4b Avian Influenza A(H5N8) Virus in Cambodia, 2021
Abstract
In late 2021, highly pathogenic avian influenza A(H5N8) clade 2.3.4.4b viruses were detected in domestic ducks in poultry markets in Cambodia. Surveillance, biosafety, and biosecurity efforts should be bolstered along the poultry value chain to limit spread and infection risk at the animal–human interface.
Since 2014, highly pathogenic avian influenza viruses (HPAIVs) with H5 hemagglutinin (HA) genes grouped in the genetic clade 2.3.4.4 have spread globally causing severe outbreaks in Africa, Europe, Asia, and most recently, North America (1). These viruses cause devastating outbreaks in poultry, rapidly evolve, and continuously reassort with other avian influenza viruses (AIVs), posing a threat to food security in many parts of the world and substantial zoonotic infection risk.
Since 2017, the Institut Pasteur du Cambodge and National Animal Health and Production Research Institute in Cambodia have partnered with the Food and Agriculture Organization of the United Nations to enhance ongoing longitudinal AIV surveillance in live bird markets and poultry storage facilities throughout Cambodia. This active surveillance reveals high levels of AIV circulation with ≈30%–50% of ducks and ≈20%–40% of chickens testing positive for various influenza A subtypes. Most detected HPAIVs were H5N1 HA clade 1 viruses during 2005–2014 and H5N1 HA clade 2.3.2.1c viruses since 2014; H5N6 clades 2.3.4.4g and 2.3.4.4h were detected sporadically during 2018–2020 (Appendix 1, Table 1). Other subtypes also circulate, including novel H7Nx low pathogenicity avian influenza viruses (LPAIVs) (2,3).
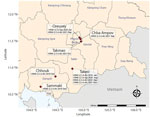
During active surveillance of live bird markets (National Ethics Committee for Health Research Approval no. 149/NECHR/2020) in late 2021, domestic ducks (Anas platyrhynchos) at Orussey (Phnom Penh, n = 1), Takmao (Kandal, n = 2), Chba Ampov (Phnom Penh, n = 1), and Takeo (Takeo, n = 1) tested positive for HPAIV H5 HA but negative for neuraminidase (NA) subtype N1 by real time reverse transcription PCR (RT-PCR). We determined these samples were the H5N8 subtype after further RT-PCR analysis (Appendix 1). Positive samples originated from Orussey and Chba Ampov markets during week 37, Takmao market during week 41, and Takeo market during week 46 of 2021 (Figure 1). Full genome sequencing on a GridION instrument (Oxford Nanopore Technologies, https://www.nanoporetech.com) confirmed these samples were HPAIV H5N8 and H5 HA clade 2.3.4.4b (4).
All H5N8 HA sequences from Cambodia encoded proteins with 2-4 amino acid differences relative to the clade 2.3.4.4b candidate vaccine strain A/Astrakhan/3212/2020(H5N8) (Table 1). HA mutations T192I and H276N (according to the H3 numbering system) were shared across all H5N8 HA proteins, whereas A188V occurred in 2 sequences, and E273K, T312S, and I339K each occurred only once. Those mutations did not correlate with previously reported phenotypic traits. Consistent with other clade 2.3.4.4b HA proteins, H5N8 viruses from Cambodia retained the HPAIV cleavage site motif, REKRRKR|GLF. The NA sequences did not contain stalk deletions or markers of antiviral drug resistance. However, other genes encoded amino acid residues associated with increased replication capacity and mammalian pathogenicity, including V89, V292, D309, R389, and T598 in PB2; G622 in PB1; D30, M43, and A215 in M1; and S42 and M106 in NS proteins (5).
We performed hemagglutination inhibition assays to assess potential cross-reactivity among the 2.3.4.4b viruses isolated from ducks in Cambodia by using 2 key reference viruses: A/Astrakhan/3212/2020, the recommended candidate vaccine virus for clade 2.3.4.4.b; and A/domestic_duck/England/074477/2021, a recently identified clade 2.3.4.4b virus from poultry associated with human infection in the United Kingdom (6). H5N8 viruses from Cambodia with V188, I192 and N276 in HA showed good recognition by antiserum raised against A/Astrakhan/3212/2020. However, A/duck/Cambodia/f1PPOreu241D3/2021 (with I192 and N276, and K339 in HA) and A/duck/Cambodia/f6T241D4/2021 (with I192, K273, N276, S312 in HA) showed reduced recognition by the antiserum (Table 2).
The H5N8 viruses from Cambodia shared >95.7% nucleotide sequence homology across their genomes and formed distinct monophyletic lineages in maximum-likelihood phylogenies of several genes (bootstrap support was 100%, except for the matrix protein gene, which was 89%; Appendix Figure 1), implying circulation of a single virus strain >10 weeks from September to November 2021. The H5 HA gene was likely derived from H5N8 viruses that have caused widespread outbreaks in poultry and wild birds across Eurasia since early 2020 (7) (Figure 2). N8 NA gene segments were closest to that of HPAIV H5N8 detected in wild and domestic waterfowl in China and Korea during 2020–21, sharing most recent ancestry with NA of A/Cygnus_columbianus/Hubei/116/2020(H5N8) that was collected in November 2020 (Appendix 1 Figure 1). Both the HA and MP gene segments were most closely related to A/brown-headed gull/Tibet/1–1/2021(H5N8), collected in May 2021 (Figure 2, panel B; Appendix 1 Figure 1). In contrast, the other gene segments encoding internal virus proteins were derived from LPAIV (Appendix 1 Figure 1). PB2 and PB1 genes shared common ancestry with LPAIV detected in ducks in Vietnam in 2020. PA and NP genes shared recent common ancestry with LPAIV isolated in 2019 from wild ducks in Korea (PA gene) and China (NP gene). The NS protein gene was most similar to that of LPAIV from ducks in China in 2018. Overall, HPAIV H5Nx clade 2.3.4.4b showed evidence of extensive genetic reassortment with LPAIV found in wild waterfowl, which frequently spillover to and from domestic poultry.
In addition to various LPAIVs, multiple H5 subtypes and clades circulate in Cambodia (Appendix 1 Table 1). H5N1 clade 2.3.2.1c viruses are detected regularly. H5N6 clade 2.3.4.4g viruses were found in Takeo and Orussey markets in chickens in 2019 and ducks in 2020, and H5N6 clade 2.3.4.4h viruses were detected sporadically in Kampot province in late 2018, Takeo province during 2019–2020, and Phnom Penh in 2020 (Figures 1, 2; Appendix 1 Figures 2–5). Therefore, detection of H5N8 clade 2.3.4.4b viruses in these same markets is a major concern because further reassortment might occur. Since 2018, outbreaks of reassorted HPAIV H5Nx clade 2.3.4.4b with NA subtypes N8, N6, N1, N3, and N5 have increased in frequency (8). These viruses have disseminated intercontinentally across migratory flyways and regionally via poultry trade, often causing considerable economic losses. In 2021, H5Nx clade 2.3.4.4b viruses caused severe outbreaks in Europe, Africa, and Asia, particularly in wild birds in western China and in domestic poultry in Vietnam (9). Since January 2022, HPAIV H5N1 clade 2.3.4.4b has been detected in waterfowl, birds of prey, and poultry across North America (10).
H5Nx clade 2.3.4.4b viruses also pose a zoonotic risk to humans and other species. In February 2021, a total of 7 cases of asymptomatic human infections with HPAIV H5N8 clade 2.3.4.4b were reported in poultry farm workers in Russia following a poultry outbreak (11). H5N6 clade 2.3.4.4 viruses have caused 79 human infection cases (including 33 cases in 2021) in China with ≈32 deaths since 2014 and 1 case in Laos (12), and 3 cases of H5Nx were reported in Nigeria (9). HPAIV H5Nx clade 2.3.4.4 viruses have also been detected in domestic cats in China and Korea (1) and red foxes in The Netherlands (1), and serologic evidence exists for infection in swine (13). More recently, HPAIV H5N1 clade 2.3.4.4b containing HA genes closely related to A/Astrakhan/3212/2020 have caused human infections in the United Kingdom (14) and United States (15). HPAIV H5N1 clade 2.3.3.4b has not been detected in Cambodia.
Because of the global spread, economic impact, and zoonotic potential of HPAIV clade 2.3.4.4b viruses, active, longitudinal surveillance in live bird markets must be maintained in Cambodia, the Greater Mekong Subregion, and globally to monitor further introduction and reassortment events. In addition, surveillance of influenza-like illness needs to be maintained among persons in close contact with infected or deceased poultry. To combat the spread of HPAIV in Cambodia and other countries, viral monitoring, biosafety, and biosecurity efforts should be bolstered along the poultry value chain. Early warning and rapid control will limit infections at the animal–human interface to reduce potential pandemic risk.
Mrs. Edwards serves as project manager for the Pathogen Evolution Laboratory in the School of Public Health at the University of Hong Kong. Her research focuses on the ecology, epidemiology, and evolution of infectious disease at the human–animal interface.
Acknowledgments
We thank persons in the Virology Unit at Institut Pasteur du Cambodge (IPC) who provided technical expertise and analysis and fruitful discussions, including Viseth Srey Horm, Songha Tok, Phalla Y, Sonita Kol, Sarath Sin, Kim Lay Chea, and Veasna Duong; the support teams at IPC, including the drivers and facilities personnel who made these studies possible; all local teams, epidemiologists, veterinary officers, and other staff from the National Animal Health and Production Research Institute; Stephanie Meyer and Tom Lewis for their critical assistance in growing viral isolates and performing hemagglutination analysis, and the authors and originating and submitting laboratories of the sequences from the GISAID database (Appendix 2).
Work at Institut Pasteur du Cambodge was supported by the Food and Agriculture Organization of the United Nations and funded by the United States Agency for International Development under the Emerging Pandemics Threats 2 (EPT-2) project. The Melbourne World Health Organization Collaborating Centre for Reference and Research on Influenza is supported by the Australian Government Department of Health. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- World Organisation for Animal Health (OIE). World animal health information system (OIE-WAHIS) [cited 2022 Sep 6]. https://wahis.oie.int
- Karlsson EA, Horm SV, Tok S, Tum S, Kalpravidh W, Claes F, et al. Avian influenza virus detection, temporality and co-infection in poultry in Cambodian border provinces, 2017-2018. Emerg Microbes Infect. 2019;8:637–9. DOIPubMedGoogle Scholar
- Vijaykrishna D, Deng YM, Grau ML, Kay M, Suttie A, Horwood PF, et al. Emergence of influenza A(H7N4) virus, Cambodia. Emerg Infect Dis. 2019;25:1988–91. DOIPubMedGoogle Scholar
- Thielen P. Influenza whole genome sequencing with integrated indexing on Oxford Nanopore platforms [cited 2022 Sep 6].
- Yamaji R, Saad MD, Davis CT, Swayne DE, Wang D, Wong FYK, et al. Pandemic potential of highly pathogenic avian influenza clade 2.3.4.4 A(H5) viruses. Rev Med Virol. 2020;30:
e2099 . DOIPubMedGoogle Scholar - Oliver I, Roberts J, Brown CS, Byrne AM, Mellon D, Hansen R, et al. A case of avian influenza A(H5N1) in England, January 2022. Euro Surveill. 2022;27:
2200061 . DOIPubMedGoogle Scholar - Lewis NS, Banyard AC, Whittard E, Karibayev T, Al Kafagi T, Chvala I, et al. Emergence and spread of novel H5N8, H5N5 and H5N1 clade 2.3.4.4 highly pathogenic avian influenza in 2020. Emerg Microbes Infect. 2021;10:148–51. DOIPubMedGoogle Scholar
- Food and Agriculture Organization of the United Nations. EMPRES-i Global Animal Disease Information System [cited 2022 Sep 6]. https://empres-i.apps.fao.org/diseases
- World Health Organization. Antigenic and genetic characteristics of zoonotic influenza A viruses and development of candidate vaccine viruses for pandemic preparedness. October 2021 [cited 2022 Sep 6]. https://apps.who.int/iris/handle/10665/347437
- U.S. Department of Agriculture Animal and Plant Health Inspection Service. 2022 detections of highly pathogenic avian influenza in wild birds [cited 2022 Mar 15]. https://www.aphis.usda.gov/aphis/ourfocus/animalhealth/animal-disease-information/avian/avian-influenza/hpai-2022/2022-hpai-wild-birds
- Pyankova OG, Susloparov IM, Moiseeva AA, Kolosova NP, Onkhonova GS, Danilenko AV, et al. Isolation of clade 2.3.4.4b A(H5N8), a highly pathogenic avian influenza virus, from a worker during an outbreak on a poultry farm, Russia, December 2020. Euro Surveill. 2021;26:
2100439 . DOIPubMedGoogle Scholar - World Health Organization. Avian influenza weekly update number 844. 2022 [cited 2022 May 22]. http://apps.who.int/iris/handle/10665/351652
- Hervé S, Schmitz A, Briand F-X, Gorin S, Quéguiner S, Niqueux É, et al. Serological evidence of backyard pig exposure to highly pathogenic avian influenza H5N8 virus during 2016–2017 epizootic in France. Pathogens. 2021;10:621. DOIPubMedGoogle Scholar
- UK Health Security Agency. Human case of avian flu detected in UK. 2022 [cited 2022 May 22]. https://www.gov.uk/government/news/human-case-of-avian-flu-detected-in-uk
- Centers for Disease Control and Prevention. Reported human infections with avian influenza A viruses. 2022 [cited 2022 May 22]. https://www.cdc.gov/flu/avianflu/reported-human-infections.htm
Figures
Tables
Cite This ArticleOriginal Publication Date: December 17, 2022
1These authors contributed equally to this article.
Table of Contents – Volume 29, Number 1—January 2023
| EID Search Options |
|---|
|
|
|
|
|
|

Please use the form below to submit correspondence to the authors or contact them at the following address:
Erik A. Karlsson, Institut Pasteur du Cambodge Virology Unit, 5 Monivong Blvd, PO Box 983, Phnom Penh, Cambodia
Top