Volume 29, Number 7—July 2023
Dispatch
Evolutionary Formation and Distribution of Puumala Virus Genome Variants, Russia
Figure 1
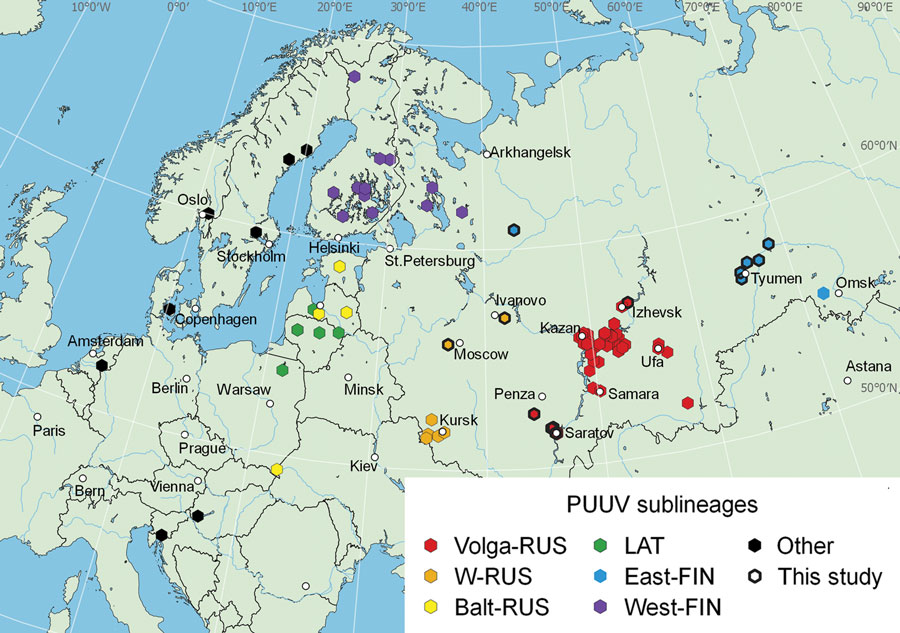
Figure 1. Locations of virus isolation in a study of the evolutionary formation and distribution of Puumala virus genome variants, Russia. The map includes all genome variants belonging to LAT, FIN (West-FIN and East-FIN sublineages), and RUS (W-RUS, Volga-RUS, and Balt-RUS) lineages that had complete coding sequences of the small (S) genome segment available in GenBank as of September 16, 2022. The map was created by using QGIS software (QGIS Development Team, http://qgis.osgeo.org). Color marking for sequences in the map correspond to those in phylogenetic trees (Figure 2). Black frames indicate isolation sites of novel sequences revealed in this study. Because other lineages were not the focus of this study, only a few Puumala virus sequences belonging to other lineages were included. An additional interactive map was generated in R (The R Foundation for Statistical Computing, https://www.r-project.org) by using the leaflet, html widgets, and webshot libraries and is available at https://rpubs.com/andreideviatkin/PUUV_RUS-FIN-LAT_lineages. Balt-RUS, sublineage from the Baltic coast region; East-FIN, sublineage from Siberia and northern Russia; FIN, Finnish lineage; LAT, Latvian lineage; RUS, Russian lineage; Volga-RUS, sublineage from the Volga River Valley; W-RUS, sublineage from western Russia; West-FIN, sublineage from Finland and Russian Karelia.