Volume 29, Number 7—July 2023
Dispatch
Evolutionary Formation and Distribution of Puumala Virus Genome Variants, Russia
Figure 2
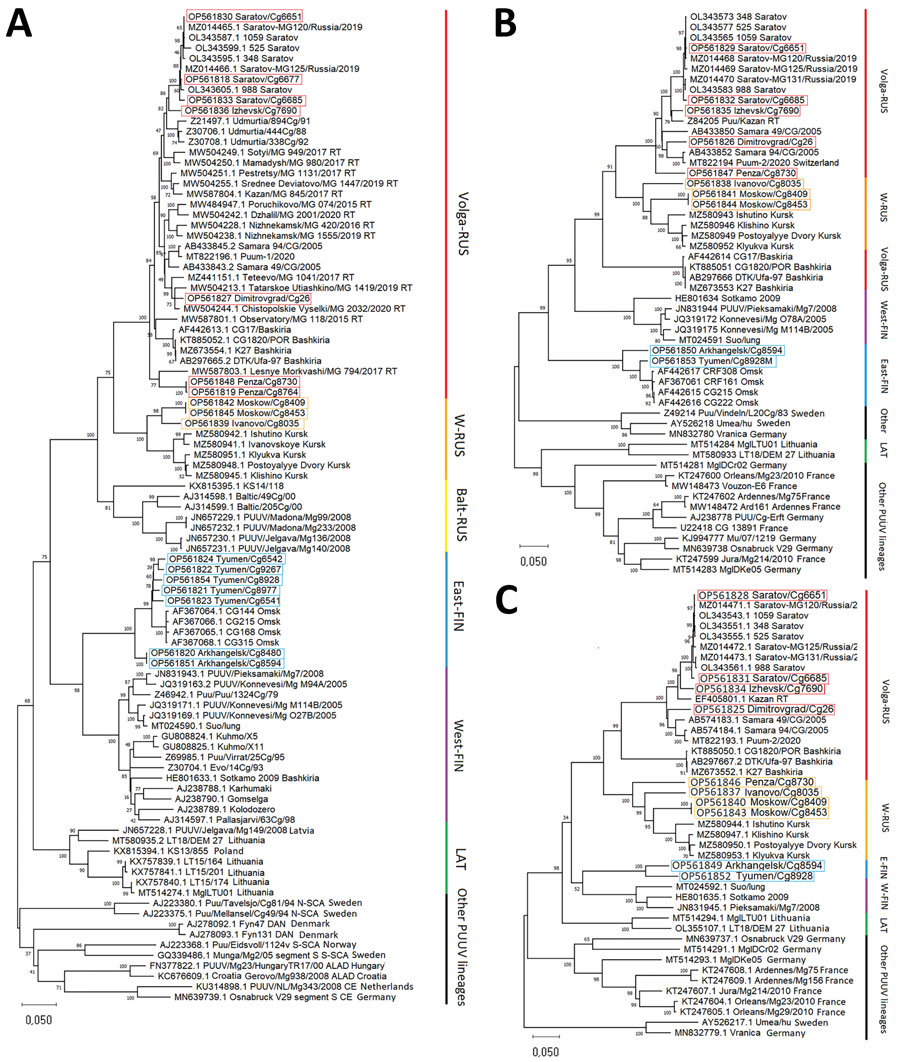
Figure 2. Phylogenetic trees of small (S), medium (M), and large (L) segments in a study of evolutionary formation and distribution of Puumala virus genome variants, Russia. A) S segment based on complete open reading frame (ORF) of 1,302 nt; B) M segment based on partial ORF 2,923 nt (525–3,447 nt of ORF of GenBank accession no. OL343565); C) L segment based on partial ORF 6,405 nt (5–6,409 nt of ORF of GenBank accession no. OL343543). The RUS lineage is divided into 3 large, color-coded subclades: Volga-RUS (red), W-RUS (orange), Balt-RUS (yellow). The FIN lineage is divided into 2 large, color-coded subclades: East-FIN (blue) and West-FIN (purple). Green indicates LAT lineage; black indicates other lineages. Boxes indicated sequences obtained in this study. GenBank accession numbers are provided for all sequences. All alignments and phylogenetic relationships of the sequences were conducted by the MUSCLE algorithm (https://www.ebi.ac.uk/Tools/msa/muscle) and maximum-likelihood method with the general time-reversible model and 1,000 bootstrap by using MEGA version X (https://www.megasoftware.net). The full S segment tree with complete dataset of all available representatives of LAT, FIN, and RUS lineages are available from https://github.com/AndreiDeviatkin/repo/blob/main/S_PUUV.png. Balt-RUS, sublineage from the Baltic coast region; East-FIN, sublineage from Siberia and northern Russia; FIN, Finnish lineage; LAT, Latvian lineage; RUS, Russian lineage; Volga-RUS, sublineage from the Volga River Valley; W-RUS, sublineage from western Russia; West-FIN, sublineage from Finland and Russian Karelia.