Volume 30, Number 3—March 2024
Synopsis
Concurrent Clade I and Clade II Monkeypox Virus Circulation, Cameroon, 1979–2022
Figure 4
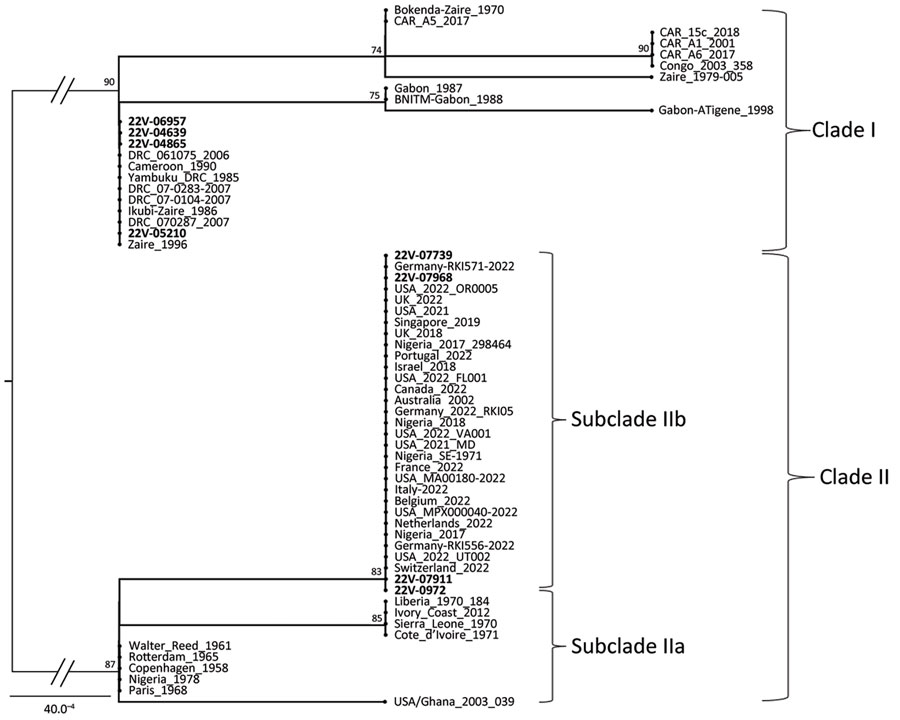
Figure 4. Maximum-likelihood phylogenetic tree of sequences in a study of concurrent clade I and clade II monkeypox virus circulation, Cameroon, 1979–2022. The tree is based on the Hasegawa-Kishino-Yano model inferred from a 942-bp fragment of the ATI gene, including 8 virus sequences from Cameroon generated in this work (bold text) and 55 reference sequences from GenBank. The tree with the highest log likelihood (−1,340.35) is shown. To test the robustness of the tree topology, 1,000 bootstrap replicates were performed. For a better display of the tree, the size of the 2 main midpoint rooted branches (represented in gray) that support the differentiation of the 2 monkeypox virus clades have been divided by half. Mpox strains from Cameroon are closely related to clades I and II, especially clade IIb for which a highlighted link to the ongoing global mpox epidemic is noted. Scale bar indicates number of substitutions per site. CAR, Central African Republic; DRC, Democratic Republic of the Congo; USA, United States.
1Current affiliation: Maryland Department of Agriculture, Salisbury, Maryland, USA.