Volume 30, Number 3—March 2024
Synopsis
Concurrent Clade I and Clade II Monkeypox Virus Circulation, Cameroon, 1979–2022
Abstract
During 1979–2022, Cameroon recorded 32 laboratory-confirmed mpox cases among 137 suspected mpox cases identified by the national surveillance network. The highest positivity rate occurred in 2022, indicating potential mpox re-emergence in Cameroon. Both clade I (n = 12) and clade II (n = 18) monkeypox virus (MPXV) were reported, a unique feature of mpox in Cameroon. The overall case-fatality ratio of 2.2% was associated with clade II. We found mpox occurred only in the forested southern part of the country, and MPXV phylogeographic structure revealed a clear geographic separation among concurrent circulating clades. Clade I originated from eastern regions close to neighboring mpox-endemic countries in Central Africa; clade II was prevalent in western regions close to West Africa. Our findings suggest that MPXV re-emerged after a 30-year lapse and might arise from different viral reservoirs unique to ecosystems in eastern and western rainforests of Cameroon.
Monkeypox virus (MPXV) is an emerging zoonotic Orthopoxvirus causing mpox in humans, a disease similar to the eradicated smallpox (1). Since identification in a monkey in 1958 (2) and a human in 1970 (3), MPXV-associated outbreaks have occurred primarily in rural rainforests in countries of Central and West Africa (4–6).
Mpox is characterized by an influenza-like syndrome accompanied by adenopathy and maculopapular rashes typically developing on the palms of the hands and soles of the feet (4,7). For infected persons, supportive care and antiviral treatments, including cidofovir and tecovirimat, are provided (4). Cross-immunity with smallpox vaccination and a new generation of smallpox vaccines equally offer some protection (8–10). However, after smallpox vaccination was discontinued in the early 1980s, herd immunity gradually declined, enabling re-emergence of mpox, which is highlighted by the increased number of cases in Africa during the past 3 decades (4,8,11–13). Since early 2022, case counts have surged, and ≈1,215 confirmed mpox cases and 219 deaths were reported in Africa by December 28, 2022 (14). Before April 2022, mpox cases in the Western Hemisphere were typically reported from exposure to the exotic pet trade and international travel (15–20). Since then, MPXV-associated outbreaks have occurred worldwide, affecting >100 countries outside Africa (4,21) and becoming a global public health concern.
Primary MPXV transmission can occur through direct contact with body fluids or skin lesions of infected animals or indirectly via contaminated fomites. Similar contact with an infected person or with infected respiratory droplets might also lead to human-to-human secondary transmission, the main transmission mode of the 2022 global outbreak (4,22). Historically, primary zoonotic transmission was more common and mostly involved an at-risk population of hunters, butchers, and bushmeat handlers; secondary transmission was rare, but nosocomial and household transmission have been described (3,13,23–25).
Phylogenetic studies report 2 distinct MPXV clades: clade I, prevalent in Central Africa, and clade II, endemic to West Africa (5,6,26–28). However, Cameroon is an exception, and both clades concurrently circulate in the country (6,29). Clade I is further subdivided into lineages 1–5 and clade II into subclades IIa and IIb; clade IIb is responsible for the multicountry outbreak that began in 2022 (27,28,30). Globally, MPXV lethality rates vary from 1% to 10%, and clade I is known to have higher mortality rates than clade II (4,24,25). The MPXV animal reservoir has not yet been identified, but the virus can infect a wide range of mammals, and Funisciurus squirrels and Graphiurus lorraineus mice are thought to be the most probable MPXV reservoirs (31–33).
In Cameroon, only 4 confirmed mpox cases were documented before the 2022 outbreak, 1 in each 1979, 1980, 1989, and 30 years later in 2018 (29,34–36). According to public health reports, more cases could have occurred and been undocumented in the country, particularly during 2018–2021, and especially in 2022, during which an mpox outbreak of unprecedented magnitude occurred and had recurrent clusters of cases (37). However, whether those infections were associated with importations from neighboring countries or from occurrence of indigenous primary or secondary transmission remains unclear (29). Overall, data on the epidemiologic features of MPXV occurrence and transmission dynamics in Cameroon are scarce. We investigated the clinical, epidemiologic, and molecular features of MPXV-associated outbreaks in Cameroon.
Sample Location
Cameroon is in central Africa and is divided into 10 administrative regions. Cameroon is known as Africa in miniature for its diverse agroecologic background: the steppe and savanna in the Far North, North, and Adamawa regions; the coastal zones in the Littoral and Southwest regions; mountain highlands in the Northwest and West regions; and the rainforest in the Centre, South, Southwest, and East regions (38). Cameroon has 3 major tropical forests: the Congo Basin Forest that extends across the East, South, and Centre regions; the Guinea moist forest in the western and Adamawa regions; and the Cameroonian Highlands forests in the Northwest and Southwest regions. Those forests are crossed by several waterways, including the Sanaga River, the largest river in Cameroon (33–40; J. Thia, master’s thesis, University of Canterbury, 2014, https://www.researchgate.net/publication/272494772_The_plight_of_trees_in_disturbed_forest_conservation_of_Montane_Trees_Nigeria).
Sample Collection
We defined a suspected case as >1 clinical signs or symptoms, including headache, asthenia, adenopathy, myalgia associated with fever, or gradually developing rashes spreading to other parts of the body, including the soles of the feet and palms of the hands. We defined a probable case as clinical manifestations without virologic confirmation but an epidemiologic link with another probable or confirmed case. A confirmed case was any case with laboratory-confirmed MPXV.
We recorded epidemiologic data, including demographic and clinical information, for all suspected cases during 1979–2022. We collected a 5-mL blood sample, vesicle swab, crust samples, or a combination of samples, from case-patients who consented to be tested. We shipped samples under a triple packaging system to the Centre Pasteur du Cameroun (CPC), which is the national reference laboratory for mpox diagnosis in Cameroon. We excluded patients from whom a sample could not be collected.
Laboratory Confirmation of MPXV Infection
At CPC, samples were received, processed, and inactivated in the Biosafety Level 3 laboratory. We purified total DNA by using the QIAamp DNA Mini Kit (QIAGEN, https://www.qiagen.com) according to the manufacturer’s instructions. We tested purified DNA for MPXV by the generic real-time PCR Taqman assay, as previously described (41). For positive samples displaying a cycle threshold (Ct) value <37, we performed further genotyping by using real-time PCRs specifically targeting MPXV clade I and II (41).
We further amplified a subset of 8 positive samples from the 2022 outbreak that had Ct values <20 by using a PCR targeting a portion of the MPXV A-type inclusion (ATI) gene, according to a previously described protocol (42). We used a 1% green-stained agarose gel to reveal resulting amplicons, which we sent to Inqaba Biotechnical Industries (Pretoria, South Africa), a commercial service provider, for Sanger sequencing.
Phylogenetical Analyses
We assembled newly determined sequences and corrected by using CLC Main Workbench software (QIAGEN). We aligned resulting consensus sequences by using MAFFT version 7 (https://mafft.cbrc.jp) and an extended dataset of 56 MPXV reference genomes from GenBank (Appendix Tables 1, 2). We submitted final alignments to the software-integrated Model Finder program (IQ-TREE, http://www.iqtree.org) to select the best evolutionary model based on Bayesian and Akaike information criterion. We used IQ-TREE version 1.6.12 (http://www.iqtree.org) to infer maximum-likelihood phylogenetic trees on MPXV ATI sequences based on the Hasegawa-Kishino-Yano plus amino acid substitution model, applying 1,000 bootstrap replicates. We submitted newly determined sequences to GenBank (accession nos. OR038717–24) (Appendix Table 2).
Statistical Analysis and Mapping
To provide a complete picture of the epidemiology of mpox in Cameroon, we added the 4 previously documented mpox cases from Cameroon to our dataset, along with available information collected from the literature and Ministry of Health archives (29,34–36). We summarized sociodemographic and clinical characteristics by using frequencies for categorical variables; we used median and interquartile range (IQR) for quantitative variables. We compared PCR-confirmed cases with nonconfirmed suspected cases by using Pearson χ2 or Fisher exact tests for categorical variables and Wilcoxon test for quantitative variables. We used univariate logistic regression to identify factors associated with MPXV infection and estimate crude odds ratios (ORs) and 95% CIs. We were unable to infer multivariable analysis models, which failed to converge because too many data were missing (Tables 1, 2). We considered p<0.05 statistically significant and p<0.07 marginally significant. We performed all analyses in R version 4.1 (The R Foundation for Statistical Computing, https://www.r-project.org). We used Quantum GIS version 3.30.1 (QGIS, https://qgis.org) to analyze and map mpox cases by health zones and geographic data.
Ethics
Sample collection and laboratory analyses were conducted within the framework of the Cameroon national surveillance program. Under that program, we obtained written or oral informed consent from all persons with suspected mpox after we provided detailed information and explanations of the sampling purpose. We obtained informed consent from parents or recognized guardians for persons <15 years of age.
Within the mpox surveillance system in Cameroon, during 1979–2022, we identified 137 suspected mpox cases, including 74 (54.41%) among male and 62 (45.59%) among female persons; 1 case had missing data for sex (Table 1). The median age of case-patients was 11 years (range 2 weeks–75 years; IQR 4–27 years); nearly half (48.18%) were <10 years of age (Table 1).
Molecular Diagnostic Results
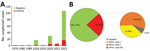
Mpox virus generic PCR showed 32 (23.36%) laboratory-confirmed mpox cases of 137 patients tested during 1979–2022 in Cameroon (Table 1; Figure 1, panel A; Appendix Table 3). Before 2018, only 3 sporadic cases were confirmed as human MPXV infection. After a 30-year gap without reported mpox cases, the surveillance system continuously identified new mpox cases during 2018–2022. Among suspected cases, only 1 was found in 2018 and 1 in 2019. In 2020 and 2021, 5 laboratory-confirmed cases were recorded each year. During 2022, mpox cases dramatically increased to 17 confirmed cases among 84 suspected cases (Figure 1, panel A; Appendix Table 3).
Genotyping of real-time PCR results identified 12 (9%) patients infected with MPXV clade I and 18 (13%) infected with MPXV clade II among 137 suspected cases; 2 (1%) historic confirmed cases lacked clade determination results (Table 1; Figure 1, panel B; Appendix Tables 2, 3). Among all laboratory-confirmed cases, only 1 death was recorded, in a patient infected with MPXV clade II. Ministry of Health investigation records indicated 2 additional patient deaths among persons with typical mpox clinical manifestations who were epidemiologically linked to 2 confirmed case-patients infected with a clade II MPXV strain. However, no specimens were collected before death; thus, we considered those probable cases. Including the probable cases, the overall case-fatality ratio (CFR) in Cameroon was 2.2% (3/139) among confirmed and suspected cases, and all deaths were associated with viral clade II.
Epidemiology and Clinical Characteristics of Confirmed Mpox Cases
Univariable analysis revealed no statistically significant difference in increased likelihood of infection by sex: 21/74 (28.38%) male and 11/62 (17.74%) female persons had confirmed MPXV infection (Table 1). MPXV-confirmed case-patients had a median age of 21.5 years (range 2 weeks–52 years; IQR 8.5–32.25 years). MPXV infection was more prevalent among adults >20 years of age; in all, 35.56% had confirmed MPXV infection, compared with 17.78% among younger MPXV-confirmed case-patients (p = 0.025). However, we saw no statistically significant difference for adults born before 1980 than for the rest of the population (p = 0.092). Larger datasets would be needed to confirm the observed trend.
MPXV infection was mostly associated with occupational activities involved in farming (OR 3.83, 95% CI 0.44–33.11) (Table 1). Similarly, potential nosocomial transmission was identified in health workers (OR 3.07, 95% CI 0.84–11.17). Other activities, including teaching, trading, or driving, when considered together, also appeared to be potential risk activities for secondary MPXV transmission (OR 2.13, 95% CI 0.52–8.77). However, we found no association for secondary transmission in the 29.82% of MPXV-confirmed cases reporting past contact with persons who had mpox-like clinical signs (Table 1). Because mpox is typically zoonotic, we also assessed antecedent of animal exposures. We observed no association with unspecified animal contacts but observed a higher risk among confirmed cases (6/13 [46.15%]) who reported contact with wild animals (OR 2.86, 95% CI 0.53–15.47) compared with persons reporting contact with domestic animals or having no contact with animals (Table 1). Among wild animal contact, study participants frequently mentioned squirrels, bats, caterpillars, pangolins, rats, porcupines, and monkeys.
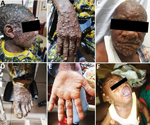
As expected from the case definition criterium requiring skin rashes, almost all (124/137 [90.5%]) MPXV-suspected cases had active skin lesions (Table 2; Figure 2; Appendix Table 3). However, we observed no specific difference for lesion progress, deepness, size, or stage among MPXV-confirmed cases compared with MPXV-negative persons (Table 2). Maculopapular lesions were more prevalent in confirmed cases who had lesions on their palms and soles (Figure 2). Clinical data identified cough (OR 2.3, 95% CI 0.95–5.59), chills or sweat (OR 3.8, 95% CI 1.52–9.48), lymphadenopathy (OR 3.53, 95% CI 1.38–9.00), sore throat when swallowing (OR 10, 95% CI 3.69–27.12), mouth ulcers (OR 8.38, 95% CI 2.8–25.09), and general fatigue (OR 3.06, 95% CI 1.17–7.98) as potential symptoms associated with MPXV infection in Cameroon (Table 2; Figure 2). Among all suspected case-patients, ≈26% who reported experiencing fever before skin rashes developed were confirmed for MPXV infection, but we saw no difference between confirmed cases with or without fever. In addition, MPXV-confirmed or -negative cases did not experience differences in headache (Table 2). We noted little difference in clinical severity in cases infected with clade I compared with those infected with clade II (Appendix Table 4). The same was true for the exposure route; we found no association between zoonotic or human-to-human transmission and a specific infecting viral clade (Appendix Table 4). However, because considerable data were missing (Tables 1, 2) we were unable to perform a multivariable analysis. Therefore, concluding interpretations of the epidemiologic and clinical features of mpox infection in Cameroon are difficult to draw.
Geographic and Phylogenetic Analysis
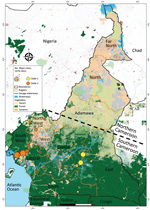
Reported suspected mpox cases originated from 8 administrative regions of Cameroon (Table 1; Figure 3). Most (97.08%) suspected cases were reported from the southern part of the country where all confirmed cases also originated. In particular, 1 (3.13%) case was confirmed in Littoral, 1 (3.13%) in the South, 3 (9.38%) in the East, 6 (18.75%) in the Northwest, 10 (31.25%) in the Southwest, and 11 (34.88%) in the Centre regions (Table 1; Figure 3; Appendix Table 3). Of note, a unique case confirmed in the Littoral region was originally from the Southwest and sought healthcare in Littoral. Genotyping of real-time PCR revealed that all clade I MPXV infections were confirmed in patients from the Centre, South, and East regions; all but 1 of clade II MPXV samples were recovered from patients from the Littoral, Northwest, and Southwest regions. Indeed, a clade II MPXV detected in the Centre region was an internally displaced person (IDP) originally from the Northwest region (Table 1; Appendix Table 3). The distribution of mpox cases points toward geographic segregation of the 2 viral clades in Cameroon. Those findings indicate a strong geographic association of MPXV genotypes in southern Cameroon, and that MPXV clade II is associated with the western part and the clade I with the eastern part of the country.
We obtained partial MPXV ATI gene sequences from 8 mpox-confirmed cases from 4 regions of Cameroon. We derived the newly determined sequences from samples collected in the Northwest (CPC code 22V-0972), Southwest (CPC codes 22V-07739, 22V-07911, 22V-07968), Centre (CPC codes 22V-05210, 22V-04865, 22V-4639), and South (CPC code 22V-6957) regions. Maximum-likelihood phylogenetic analysis of the 942 nt consensus sequences, including reference sequences (Appendix Tables 1–3), revealed that the 8 MPXV genomes from Cameroon segregated into clade I and clade II. As expected from the geographic association of MPXV isolates we report, MPXV clade I from the Centre and South regions grouped reliably with reference counterparts previously reported from countries in Central Africa, and clade II sequences from the Northwest and Southwest regions grouped consistently with strains from West Africa (Figure 4). Clade II strains from Cameroon clustered reliably within subclade IIb with 83% bootstrap support (Figure 4). Altogether, genotypic and phylogenetic analysis confirmed the concurrent circulation of both MPXV clades I and II in Cameroon with a striking geographic segregation.
We examined the clinical, epidemiologic, and molecular patterns of MPXV infection in Cameroon over a 44-year period (1979–2022) as part of mpox surveillance in the country. During 1979–2022, a total of 137 persons were suspected of having mpox, and 32 were confirmed to be MPXV infected. Three persons died (CFR 2.2%) and death was associated with MPXV clade II. That CRF is much lower than those reported in previous studies of MPXV clade I that showed CFRs of 7%–10% (13,43). Overall, CFRs are lower among patients infected with clade II, including in the 2022 global outbreak settings (4,25). We were not able to collect information on potential underlying conditions of case-patients to determine whether immunocompromising conditions contributed to death, which would have worsened the clinical disease manifestations, as highlighted by others (44). In addition, fatal cases associated with clade I potentially escaped the national surveillance system in Cameroon, which is new and still being improved.
We found that both primary zoonotic and secondary human-to-human MPXV transmission occurs in Cameroon, including nosocomial transmission affecting health workers. Our results are consistent with reports describing secondary transmission chains, including intrafamilial transmission and occupational transmission through trade, transportation, hunting, and healthcare in endemic countries (24,43,45,46). This study highlights a common MPXV acquisition pathway in endemic countries, interspecies transmission, and wild animals are presumed reservoirs of the virus (31,32,47). Distinguishing between primary and secondary transmission is difficult because both could occur. Additional data and further investigations are required to clearly understand the underlying drivers of MPXV transmission in Cameroon.
A limitation of this study is our inability to perform more precise analyses to determine the characteristics independently describing the mpox epidemiology in Cameroon. Because the current surveillance system is still handwritten and forms are often incompletely filled, data are missing, as is common in paper-based data collection systems (48).
Since 1979, MPXV infections in Cameroon have occurred in 6 of the 10 administrative divisions of the country: Centre, South, East, Littoral, Northwest, and Southwest. All those administrative divisions are in the southern part of the country, which is a forested area encompassed by the lower montane forest of Guinea and the tropical rainforest of the Congo Basin, a favorable ecosystem for potential wildlife hosts. In contrast, northern Cameroon, a dry Sahelian and savannah zone, seems unlikely to be conducive to MPXV transmission because no cases have been confirmed in this region. That ecosystem is probably not suitable for MPXV reservoirs due to the dry environment. In most endemic countries, including Sierra Leone, Nigeria, Liberia, Central African Republic, and the Democratic Republic of the Congo, mpox cases mainly have been reported from forested areas (24,25,46). Most MPXV-confirmed cases in our study originated from the Centre (34 [38%]) and Southwest (31 [25%]) regions, which are the 2 most affected areas in the country. The Northwest region was the third (18 [75%] cases) most affected region. The Northwest and Southwest regions have been most seriously affected by civil unrest since 2017. That civil unrest has increased the number of IDPs in the country, and IDPs often move to different regions and neighboring countries. Furthermore, that situation has greatly increased human contact with wildlife as IDPs seek refuge in makeshift camps in the forest. By living in overlapping natural habitats of wild animals and potential MPXV reservoirs, populations of the Southwest and Northwest regions are under increased threat of zoonotic MPXV acquisition. Indeed, in Africa, civil unrest often leads to increases in mpox cases, and risk for any zoonotic disease is common (4,49). In several endemic countries, mpox outbreaks in the context of armed conflicts or massive population movements are a typical epidemiologic feature, and those conditions are usually associated with inefficient disease surveillance and control (4,49).
Genotypic and phylogenetic analyses revealed that both clade I and clade II are concurrently circulating in Cameroon and that a clear geographic segregation appears between the 2 clades. Circulation of both MPXV clades in Cameroon was previously reported in 2 published MPXV sequences from Cameroon (6,29). However, this study builds on those findings and provides more samples to further confirm that clades I and II concurrently circulate in a single country, a unique feature in MPXV epidemiology.
The geographic segregation of the clades is more perceptible in clade II case 21V-04877 in the Centre region. An epidemiologic investigation revealed that the case-patient was an IDP originating from the Northwest region, where MPXV clade II is endemic. The geographic segregation observed between MPXV strains circulating in Cameroon can be attributed to the natural barriers that potential animal reservoirs might not be able to cross between the Centre, East, and South regions, covered by the Congo Basin tropical forest, and the Northwest and Southwest regions, covered by lower montane moist forest of Guinea (38,40). Indeed, the Sanaga River, which is the largest river in the country, and the Cameroon highlands region sharply separate the 2 geographic areas into tropical moist forest ecoregions. The Cross-Sanaga-Bioko coastal forests lie to the north between the Sanaga River and the Cross River of Nigeria, and the Atlantic Equatorial coastal forests extends south of the river through southwestern Cameroon and other neighboring countries of central Africa (38,39). Alternatively, the 2 ecologic environments potentially host different reservoirs. Several studies aimed to identify presumed MPXV reservoirs (31,33,47), but none have emphasized the potential of 2 distinct reservoirs that could be specific to a given ecosystem. Furthermore, MPXV circulation in humans in Cameroon after decades of absence might have resulted from movements of human populations, reservoir hosts, or both from endemic reservoirs in neighboring countries as armed conflicts intensified cross-border movements since 2017. That hypothesis is supported by the clustering of newly sequenced MPXV strains with counterparts originating from neighboring countries that have no physical barrier with the eastern and western parts of Cameroon but have long terrestrial borders.
In summary, this study provides detailed insight into the mpox epidemic in Cameroon during a 44-year period. The epidemiology of mpox in Cameroon involves both primary and secondary transmission. Segregated clade I and II virus strains concurrently circulate, suggesting potential existence of distinct viral reservoirs and cross-border circulation of MPXV. This study can inform the design, optimization, and evaluation of public health interventions for monitoring and controlling mpox in Cameroon and other countries in Africa with similar epidemiologic settings.
Dr. Djuicy is a research scientist working at the virology department at the Centre Pasteur du Cameroon in Yaoundé, on zoonosis and emerging diseases including viral hemorrhagic fevers. Her research interests focus on developing research axes for emerging and reemerging neglected and poverty-related viral diseases, including mpox, Ebola, Lassa Fever, and Marburg virus.
Acknowledgments
We thank Africa Centres for Disease Control and Prevention for mpox diagnostic reagents provided to the surveillance system. We thank the patients or their legal guardians who agreed to be enrolled in this study. We thank all the personnel who identified and collected samples from suspected cases included in this study. We thank Huguette Simo for technical review of the manuscript, Landry Messanga for inference of the raw phylogenetic tree, and Hornela Ossombo for QGIS mapping.
This work was supported by the Centre Pasteur of Cameroon.
References
- Marennikova SS, Moyer RW. Classification of poxviruses and brief characterization of the genus Orthopoxvirus. In: Shchelkunov SN, Marennikova SS, Moyer RW, editors. Orthopoxviruses pathogenic for humans. Boston: Springer; 2005. p. 11–8.
- von Magnus P, Andersen EK, Petersen KB, Birch-Andersen A. A pox-like disease in cynomolgus monkeys. Acta Pathol Microbiol Scand. 1959;46:156–76. DOIGoogle Scholar
- Ladnyj ID, Ziegler P, Kima E. A human infection caused by monkeypox virus in Basankusu Territory, Democratic Republic of the Congo. Bull World Health Organ. 1972;46:593–7.PubMedGoogle Scholar
- Gessain A, Nakoune E, Yazdanpanah Y. Monkeypox. N Engl J Med. 2022;387:1783–93. DOIPubMedGoogle Scholar
- Faye O, Pratt CB, Faye M, Fall G, Chitty JA, Diagne MM, et al. Genomic characterisation of human monkeypox virus in Nigeria. Lancet Infect Dis. 2018;18:246. DOIPubMedGoogle Scholar
- Nakazawa Y, Mauldin MR, Emerson GL, Reynolds MG, Lash RR, Gao J, et al. A phylogeographic investigation of African monkeypox. Viruses. 2015;7:2168–84. DOIPubMedGoogle Scholar
- Rimoin AW, Mulembakani PM, Johnston SC, Lloyd Smith JO, Kisalu NK, Kinkela TL, et al. Major increase in human monkeypox incidence 30 years after smallpox vaccination campaigns cease in the Democratic Republic of Congo. Proc Natl Acad Sci U S A. 2010;107:16262–7. DOIPubMedGoogle Scholar
- Rao AK, Petersen BW, Whitehill F, Razeq JH, Isaacs SN, Merchlinsky MJ, et al. Use of JYNNEOS (smallpox and monkeypox vaccine, live, nonreplicating) for preexposure vaccination of persons at risk for occupational exposure to orthopoxviruses: recommendations of the Advisory Committee on Immunization Practices—United States, 2022. MMWR Morb Mortal Wkly Rep. 2022;71:734–42. DOIPubMedGoogle Scholar
- Eto A, Saito T, Yokote H, Kurane I, Kanatani Y. Recent advances in the study of live attenuated cell-cultured smallpox vaccine LC16m8. Vaccine. 2015;33:6106–11. DOIPubMedGoogle Scholar
- Reynolds MG, Damon IK. Outbreaks of human monkeypox after cessation of smallpox vaccination. Trends Microbiol. 2012;20:80–7. DOIPubMedGoogle Scholar
- Simpson K, Heymann D, Brown CS, Edmunds WJ, Elsgaard J, Fine P, et al. Human monkeypox - After 40 years, an unintended consequence of smallpox eradication. Vaccine. 2020;38:5077–81. DOIPubMedGoogle Scholar
- Besombes C, Mbrenga F, Schaeffer L, Malaka C, Gonofio E, Landier J, et al. National monkeypox surveillance, Central African Republic, 2001–2021. Emerg Infect Dis. 2022;28:2435–45. DOIPubMedGoogle Scholar
- Africa Centres for Disease Control and Prevention. Outbreak brief 24: mpox in Africa Union member states [cited 2023 Nov 23]. https://africacdc.org/disease-outbreak/outbreak-brief-24-mpox-in-africa-union-member-states
- Erez N, Achdout H, Milrot E, Schwartz Y, Wiener-Well Y, Paran N, et al. Diagnosis of imported monkeypox, Israel, 2018. Emerg Infect Dis. 2019;25:980–3. DOIPubMedGoogle Scholar
- Ng OT, Lee V, Marimuthu K, Vasoo S, Chan G, Lin RTP, et al. A case of imported Monkeypox in Singapore. Lancet Infect Dis. 2019;19:1166. DOIPubMedGoogle Scholar
- Vaughan A, Aarons E, Astbury J, Brooks T, Chand M, Flegg P, et al. Human-to-human transmission of monkeypox virus, United Kingdom, October 2018. Emerg Infect Dis. 2020;26:782–5. DOIPubMedGoogle Scholar
- Vaughan A, Aarons E, Astbury J, Balasegaram S, Beadsworth M, Beck CR, et al. Two cases of monkeypox imported to the United Kingdom, September 2018. Euro Surveill. 2018;23:
1800509 . DOIPubMedGoogle Scholar - Yong SEF, Ng OT, Ho ZJM, Mak TM, Marimuthu K, Vasoo S, et al. Imported Monkeypox, Singapore. Emerg Infect Dis. 2020;26:1826–30. DOIPubMedGoogle Scholar
- Reed KD, Melski JW, Graham MB, Regnery RL, Sotir MJ, Wegner MV, et al. The detection of monkeypox in humans in the Western Hemisphere. N Engl J Med. 2004;350:342–50. DOIPubMedGoogle Scholar
- Centers for Disease Control and Prevention. 2022–2023 Mpox outbreak global map [cited 2023 May 9]. https://www.cdc.gov/poxvirus/mpox/response/2022/world-map.html
- Kaler J, Hussain A, Flores G, Kheiri S, Desrosiers D. Monkeypox: a comprehensive review of transmission, pathogenesis, and manifestation. Cureus. 2022;14:
e26531 . DOIPubMedGoogle Scholar - Quiner CA, Moses C, Monroe BP, Nakazawa Y, Doty JB, Hughes CM, et al. Presumptive risk factors for monkeypox in rural communities in the Democratic Republic of the Congo. PLoS One. 2017;12:
e0168664 . DOIPubMedGoogle Scholar - Nakoune E, Lampaert E, Ndjapou SG, Janssens C, Zuniga I, Van Herp M, et al. A nosocomial outbreak of human monkeypox in the Central African Republic. Open Forum Infect Dis. 2017;4:ofx168.
- Yinka-Ogunleye A, Aruna O, Dalhat M, Ogoina D, McCollum A, Disu Y, et al.; CDC Monkeypox Outbreak Team. Outbreak of human monkeypox in Nigeria in 2017-18: a clinical and epidemiological report. Lancet Infect Dis. 2019;19:872–9. DOIPubMedGoogle Scholar
- Likos AM, Sammons SA, Olson VA, Frace AM, Li Y, Olsen-Rasmussen M, et al. A tale of two clades: monkeypox viruses. J Gen Virol. 2005;86:2661–72. DOIPubMedGoogle Scholar
- Happi C, Adetifa I, Mbala P, Njouom R, Nakoune E, Happi A, et al. Urgent need for a non-discriminatory and non-stigmatizing nomenclature for monkeypox virus. PLoS Biol. 2022;20:
e3001769 . DOIPubMedGoogle Scholar - Isidro J, Borges V, Pinto M, Sobral D, Santos JD, Nunes A, et al. Phylogenomic characterization and signs of microevolution in the 2022 multi-country outbreak of monkeypox virus. Nat Med. 2022;28:1569–72. DOIPubMedGoogle Scholar
- Sadeuh-Mba SA, Yonga MG, Els M, Batejat C, Eyangoh S, Caro V, et al. Monkeypox virus phylogenetic similarities between a human case detected in Cameroon in 2018 and the 2017-2018 outbreak in Nigeria. Infect Genet Evol. 2019;69:8–11. DOIPubMedGoogle Scholar
- Berthet N, Descorps-Declère S, Besombes C, Curaudeau M, Nkili Meyong AA, Selekon B, et al. Genomic history of human monkey pox infections in the Central African Republic between 2001 and 2018. Sci Rep. 2021;11:13085. DOIPubMedGoogle Scholar
- Khodakevich L, Jezek Z, Kinzanzka K. Isolation of monkeypox virus from wild squirrel infected in nature. Lancet. 1986;1:98–9. DOIPubMedGoogle Scholar
- Radonić A, Metzger S, Dabrowski PW, Couacy-Hymann E, Schuenadel L, Kurth A, et al. Fatal monkeypox in wild-living sooty mangabey, Côte d’Ivoire, 2012. Emerg Infect Dis. 2014;20:1009–11. DOIPubMedGoogle Scholar
- Curaudeau M, Besombes C, Nakouné E, Fontanet A, Gessain A, Hassanin A. Identifying the most probable mammal reservoir hosts for monkeypox virus based on ecological niche comparisons. Viruses. 2023;15:727. DOIPubMedGoogle Scholar
- Tchokoteu PF, Kago I, Tetanye E, Ndoumbe P, Pignon D, Mbede J. [Variola or a severe case of varicella? A case of human variola due to monkeypox virus in a child from the Cameroon] [in French]. Ann Soc Belg Med Trop. 1991;71:123–8.PubMedGoogle Scholar
- Eozenou P. Retrospective investigation into a case of monkeypox in the United Republic of Cameroon [in French]. Bull OCEAC. 1980;2:23–6.
- Heymann D. Initial report of an Ebola-Monkeypox investigation in Moloundou (Cameroon, February 1980) [in French]. Bull OCEAC. 1980;7:58–60.
- Public Health Emergency Operations Coordination Center. Cameroon. Monkeypox [in French] [cited 2023 May 9]. https://www.ccousp.cm/urgences-sanitaires/Mpox/situation-Mpox-cameroun
- Molua EL, Lambi CM. Climate, hydrology and water resources in Cameroon. Pretoria: The Centre for Environmental Economics and Policy in Africa (CEEPA), University of Pretoria South Africa; 2006.
- Zébazé Togouet SH, Nyamsi Tchatcho N, Tharme RE, Piscart C. The Sanaga River, an example of biophysical and socio-cultural integration in Cameroon, Central Africa. In: Wantzen KM, editor. River culture: life as a dance to the rhythm of the waters. Paris: United Nations Educational, Scientific and Cultural Organization; 2023.
- One Earth. Afrotropics [cited 2023 May 9]. https://www.oneearth.org/realms/afrotropics
- Li Y, Zhao H, Wilkins K, Hughes C, Damon IK. Real-time PCR assays for the specific detection of monkeypox virus West African and Congo Basin strain DNA. J Virol Methods. 2010;169:223–7. DOIPubMedGoogle Scholar
- Meyer H, Ropp SL, Esposito JJ. Gene for A-type inclusion body protein is useful for a polymerase chain reaction assay to differentiate orthopoxviruses. J Virol Methods. 1997;64:217–21. DOIPubMedGoogle Scholar
- Nolen LD, Osadebe L, Katomba J, Likofata J, Mukadi D, Monroe B, et al. Extended human-to-human transmission during a monkeypox outbreak in the Democratic Republic of the Congo. Emerg Infect Dis. 2016;22:1014–21. DOIPubMedGoogle Scholar
- Ogoina D, Iroezindu M, James HI, Oladokun R, Yinka-Ogunleye A, Wakama P, et al. Clinical course and outcome of human monkeypox in Nigeria. Clin Infect Dis. 2020;71:e210–4. DOIPubMedGoogle Scholar
- Larway LZ, Amo-Addae M, Bulage L, Adewuyi P, Shannon F, Wilson W, et al. An outbreak of monkeypox in Doedain District, Rivercess County, Liberia, June, 2017. J Interv Epidemiol Public Heal. 2021;4:8. DOIGoogle Scholar
- Besombes C, Gonofio E, Konamna X, Selekon B, Grant R, Gessain A, et al. Intrafamily transmission of monkeypox virus, Central African Republic, 2018. Emerg Infect Dis. 2019;25:1602–4. DOIPubMedGoogle Scholar
- Reynolds MG, Doty JB, McCollum AM, Olson VA, Nakazawa Y. Monkeypox re-emergence in Africa: a call to expand the concept and practice of One Health. Expert Rev Anti Infect Ther. 2019;17:129–39. DOIPubMedGoogle Scholar
- Tchatchueng-Mbougua JB, Messanga Essengue LL, Septoh Yuya FJ, Kamtchogom V, Hamadou A, Sadeuh-Mbah SA, et al. Improving the management and security of COVID 19 diagnostic test data with a digital platform in resource-limited settings: The case of PlaCARD in Cameroon. PLOS Digit Health. 2022;1:
e0000113 . DOIPubMedGoogle Scholar - McPake B, Witter S, Ssali S, Wurie H, Namakula J, Ssengooba F. Ebola in the context of conflict affected states and health systems: case studies of Northern Uganda and Sierra Leone. Confl Health. 2015;9:23. DOIPubMedGoogle Scholar
Figures
Tables
Cite This ArticleOriginal Publication Date: February 07, 2024
1Current affiliation: Maryland Department of Agriculture, Salisbury, Maryland, USA.
Table of Contents – Volume 30, Number 3—March 2024
| EID Search Options |
|---|
|
|
|
|
|
|

Please use the form below to submit correspondence to the authors or contact them at the following address:
Richard Njouom, Centre Pasteur du Cameroun, PO Box 1274, Yaounde, Cameroon
Top