Volume 24, Number 2—February 2018
Research
Yersinia pestis Survival and Replication in Potential Ameba Reservoir
Abstract
Plague ecology is characterized by sporadic epizootics, then periods of dormancy. Building evidence suggests environmentally ubiquitous amebae act as feral macrophages and hosts to many intracellular pathogens. We conducted environmental genetic surveys and laboratory co-culture infection experiments to assess whether plague bacteria were resistant to digestion by 5 environmental ameba species. First, we demonstrated that Yersinia pestis is resistant or transiently resistant to various ameba species. Second, we showed that Y. pestis survives and replicates intracellularly within Dictyostelium discoideum amebae for ˃48 hours postinfection, whereas control bacteria were destroyed in <1 hour. Finally, we found that Y. pestis resides within ameba structures synonymous with those found in infected human macrophages, for which Y. pestis is a competent pathogen. Evidence supporting amebae as potential plague reservoirs stresses the importance of recognizing pathogen-harboring amebae as threats to public health, agriculture, conservation, and biodefense.
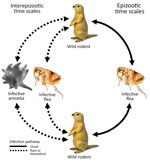
The etiologic agent of plague, Yersinia pestis, is a gram-negative coccobacillus and a facultative intracellular pathogen. Y. pestis exhibited the highest overall mortality rate of any infectious disease from its earliest recorded emergence through 1941 (1). During 2010–2015, a mean of 650 cases were reported globally each year, with a case fatality rate of 23%–41% (depending on manifestation as bubonic, pneumonic, or septicemic plague), rising to 66%–100% when adequate medical care was not promptly received (2). Y. pestis primarily infects small ground-dwelling mammals, specifically of the taxonomic order Rodentia, but maintains high spillover potential to other vertebrates, including humans, caused by its high virulence and fleaborne transmission. Epizootic plague is typically vectored by multiple flea species and is transmitted within and between meta-populations of hosts by flea bites (Figure 1).
Plague ecology is characterized by sporadic epizootics, followed by 2–5-year cryptic dormancy periods (3–9). Despite much information on epizootic transmission mechanisms, little is known about the origin of re-emergent plague cases in wild animal populations (Figure 1). Plague among wild animals commonly re-emerges in plague foci after multiple years of inactivity, despite ongoing biosurveillance and attempts at detection during interepizootic periods. The existence of environmental plague reservoirs has been theorized for >80 years (3–13). Various avenues of recent research suggest that soil-dwelling amebae may be competent environmental reservoirs of Y. pestis. Amebae are a taxonomically diverse group of phagocytic organisms residing in every major lineage of eukaryotes. Amebae are pervasive in soil and water environments and are recognized for their ability to harbor pathogens that drastically affect ecologic communities (14–19). Free-living amebae cycle between 2 distinct life-states: trophozoites, an active, mobile, feeding state; and cysts or spores, a robust dormant state induced in part by adverse environmental conditions.
Ameba reservoir potential for Y. pestis is indicated by 4 major factors: the ability of related Y. enterocolitica and Y. pseudotuberculosis bacteria to persist in protozoan amebae (20–22); correlative data indicating plague epizootics temporally follow periods of increased precipitation known to reanimate ameba cysts (5,23,24); the demonstrated ability of Y. pestis to express various proteins enabling escape of the phagolysosome in a diverse array of phagocytic cells including human macrophages (25–27); and prior associations between Y. pestis and the soil amebae, Vermamoeba (formerly Hartmanella) rhysodes and Acanthamoeba castellanii, that demonstrate intracellular persistence up to 5 days (13,28,29). Amebae display a high degree of functional homology with mammalian macrophages, leading to the description of amebae as feral macrophages. The ameba reservoir hypothesis is compelling for many pathogens with unexplained sporadic occurrence and cryptic dormancy periods as supported by a growing catalog (>225) of intracellular pathogens capable of surviving and/or replicating within amebae under diverse conditions (14,17,18,30).
We tested the hypothesis that 5 species of environmentally ubiquitous amebae demonstrate reservoir potential for the maintenance of Y. pestis. We implemented field and laboratory investigations to assess environmental co-occurrence of study ameba species with plague epizootics; experimental infection prevalence in amebae; experimental infection intensity; intraameba bacterial location; bacterial viability postphagocytosis; and bacterial replication inside trophozoite amebae. We discuss the potential for D. discoideum ameba to act as interepizootic reservoirs, the functional homology between phagocytic amebae and mammalian macrophages, and the ability of ameba to exert selective pressure on the evolutionary trajectory of pathogen virulence and transmission mode. Further, we stress the importance of recognizing pathogen-harboring amebae as potential threats to global health, agriculture, conservation, and biodefense.
By using field experiments, we molecularly assessed the co-occurrence of amebae and Y. pestis in prairie dog burrows in the Pawnee National Grassland of northeastern Colorado, USA. This grassland is an established plague foci that has exhibited recurrent plague epizootics since ≈1940 (31). We used molecular analyses of soil and amebae cultured from the soil to identify candidate ameba species that may act as reservoirs for plague persistence.
Plague-Endemic Soil Isolates
We selected 24 prairie dog burrows from 8 prairie dog colonies, which can contain hundreds of animals, on the basis of suspected plague presence indicated by sustained decreases in population size during a 3-week observation period in August 2016 (Technical Appendix Figure 1). We selected individual burrows within the colony boundaries on the basis of apparent prairie dog activity (feces, freshly excavated soil, and noncollapsed burrow structure) and along a gradient from the center of the colony to the periphery. We collected soil by attaching 50-mL conical tubes to a 6-m flexible metal probe, maneuvering the probe into the prairie dog burrow to maximum achievable depth, and using the probe to scrape soil into the tubes. We sealed viable soil samples (>20 mL from >3 m deep) and stored them at 22°C until processing within 12 hours.
Cultivation of Amebae from Soil
We isolated amebae from soil in plague-affected prairie dog burrows by using modified culture methods (32) (Technical Appendix Figure 2), incubated culture plates at 28°C, and observed for changes daily. We supplemented liquid medium with gentamicin (200 μg/mL) after 72 hours or at earliest detection of any bacterial growth. We aseptically transferred ameba cultures without bacterial contamination to 25-cm2 tissue culture flasks in ameba-specific media containing penicillin/streptomycin. We identified ameba by using multiplex and endpoint PCR after extracting DNA by using a QIAGEN DNeasy Blood & Tissue Kit ( QIAGEN, Hilden, Germany) (33,34) (Technical Appendix Figure 3).
Bacterial Strains and Culture Conditions
We cultured Y. pestis strains from frozen stocks in lysogeny broth (LB) medium. We used 2 strains of Y. pestis throughout the study: a nontransformed prototypical strain of Y. pestis CO92 and a recombinant gfp-expressing strain, Y. pestis CO92 pgm+, pCD1, pGFPuv, amp+, from the Centers for Disease Control and Prevention (Fort Collins, CO, USA). We cultured the transformed strain by using 100 μg/mL carbenicillin to maintain selective pressure for retention of gfp plasmids. Culture conditions simulated a mammalian host environment (37°C for 24 h to stationary phase) and then an extra-host environment (28°C for 24 h) to activate phenotypically plastic expression profiles. We monitored bacterial growth spectrophotometrically at OD600.
Ameba Strains and Culture Conditions
We obtained stocks of A. lenticulata (ATCC 30841), A. castellanii (ATCC 30234), A. polyphaga Linc-Ap1 (CCAP 1501/18), and V. vermiformis (ATCC 50237) from the American Type Culture Collection (Manassas, VA, USA) and the Culture Collection of Algae and Protozoa (https://www.ccap.ac.uk/) and Dictyostelium discoideum (NC4A2) from DictyBase (http://dictybase.org/). We axenically cultivated ameba stocks with genera-specific media in T25 tissue culture flasks at 28°C and verified them to be axenic by using standardized methods (19,35–37).
Co-culture Experiments
Intraameba Infection Prevalence and Intensity Assays
We individually co-cultured laboratory ameba species with Y. pestis by using established methods (21). We adjusted viable ameba trophozoite densities to 5 × 105 trophozoites/mL in triplicate 25-cm2 tissue culture flasks and combined Y. pestis (CO92 pgm+, pCD1, pGFPuv, amp+) cultures with ameba flasks (excluding ameba controls), resulting in 5 × 107 viable Y. pestis cells/mL and a multiplicity of infection (MOI) of 100 on the basis of OD 600 calculations. We incubated co-cultures at 28°C for 4 h before removing infected amebae, ameba controls, and bacteria controls from the surface of the flasks and washing them 3 times with Page amoeba saline (PAS) at 100 × g for 5 min (36). We then exposed amebae to gentamicin (100 μg/mL) for either 1 or 4 h to eliminate residual extracellular bacteria, then washed them 3 more times to remove antibiotic drug residue. Finally, we removed the supernatant from the last wash, concentrated it via centrifugation (4,400 × g for 10 min), then plated it on LB agar to ensure no extracellular bacteria persisted.
We fixed aliquots of each infected ameba treatment in 4% paraformaldehyde for 15 min before washing (4,400 × g, 5 min) and resuspending in 1× PAS for microscopic analysis. We used a fluorescent confocal microscope (Zeiss LSM 510 with ZEN 2009 SP2 software [Carl Zeiss GmbH, Thornwood, NY, USA]) to determine mean infection prevalence (the percentage of amebae containing >1 intracellular Y. pestis bacterium across 16 fields of view per treatment replicate). We determined mean infection intensity and its distribution by quantifying the number of intracellular bacteria per individual infected ameba, verified by z-stack confocal microscopy across 16 fields of view per treatment replicate. We used 1-way measured analysis of variance (ANOVA) on prevalence and intensity means across all 5 amebae species. We log-transformed data as necessary to meet model assumption and used least-squared mean analyses with Tukey’s adjustments for pairwise comparisons.
Ultrastructural Description of Intraameba Bacterial Location
We used Y. pestis (CO92 pgm+, pCD1, pGFPuv, amp+) in similar co-culture infection assays with A. castellanii (MOI 100 in PAS at 28°C). We co-cultured bacteria for durations of 10 min, 30 min, and 24 h to assess proximal and delayed effects of phagocytosis on bacterial cell viability and intraameba bacterial location. After co-culture, mixtures were fixed in standard electron microscopy fixative for 2 h, then washed 2 times in 0.1 M cacodylate buffer. We then shipped fixed samples in 0.1 M cacodylate buffer to the Cryo-electron Microscopy Laboratory at the University of Texas Medical Branch (Galveston, TX, USA) for transmission electron microscopy (TEM) according to standardized procedures. We determined bacterial location within amebae by ultrastructural analysis of transmission electron micrographs and studied intracellular bacterial morphology to assess ameba-mediated bacterial lysis as measured by cell size, shape, and apparent membrane integrity.
Intra-meba Bacterial Survival and Quantification of Intraameba Bacterial Replication
We performed intraameba bacterial survival and replication assays in triplicate across 5 ameba species (A. castellanii, A. lenticulata, A. polyphaga, D. discoideum, and V. vermiformis); 2 bacteria species (Y. pestis CO92 and Escherichia coli); 5 postinfection time points (0, 1, 4, 24, and 48 h); and 3 antibiotic drug exposure periods (0, 1, and 4 h) for removing extracellular bacteria postinfection. We used E. coli as an ameba-susceptible control bacterium. We performed co-cultures in 200-μL volumes within 96-well plates at a MOI of 100 in 1/2× dilute ameba growth medium at 28°C for 1 h and used control ameba and bacteria as monocultures. After initial infection, we removed the supernatant of each well, washed wells 3 times with PAS, exposed them to gentamicin (100 μg/mL), washed 3 times more, and incubated them in PAS. PAS was verified to be bacteriostatic to Y. pestis, thereby precluding extracellular bacterial replication and continuous ingestion by amebae. We lysed infected ameba trophozoites selectively with 100 μL 0.5% sodium deoxycholate for 5 min before serially diluting and plating on LB agar. We incubated plates at 28°C for 48 h before counting CFUs to determine bacterial survival and replication after phagocytosis by amebae. The 0.5% sodium deoxycholate lysing detergent had no effect on CFU counts in bacterial monoculture controls (data not shown).
Y. pestis and 5 species of amebae co-occur in soils of prairie dog burrows undergoing plague epizootics. We cultured a wide diversity of amebae from soil within plague-positive prairie dog burrows in northeastern Colorado and identified live amebae of each study species (Technical Appendix Figure 3). Among 8 prairie dog colonies, 24 burrows sampled yielded 15 heterogeneous ameba cultures free of bacteria or fungi. We identified >1 Acanthamoeba spp. From 86.6% of cultures (n = 13), D. discoideum from 53.3% of cultures (n = 8), and V. vermiformis from 6.6% of cultures (n = 1).
Y. pestis is phagocytosed by amebae with heterogeneous prevalence and intensity. Representative fluorescent confocal micrographs of A. castellanii and D. discoideum illustrate differences in infection intensity and prevalence (Figure 2). ANOVA F-test results indicate significant differences in infection prevalence (or phagocytic efficiency) among ameba species (p = 0.0231) (Table). Repeat experiments maintained relative ranking of mean infection intensity and infection prevalence across ameba species (A. castellanii, n = 1,441; A. lenticulata, n = 1,156; A. polyphaga, n = 737; D. discoideum, n = 624; and V. vermiformis, n = 528). Pairwise comparisons indicate V. vermiformis has significantly lower infection prevalence than A. lenticulata (p = 0.0344). Infection prevalence ranged from 24.07% in 1 replicate of V. vermiformis to 54.83% in 1 replicate of A. lenticulata.
Infection intensity was also significantly different among ameba species (p = 0.0014) (Table). Pairwise comparisons showed V. vermiformis has a significantly lower infection intensity than both A. lenticulata (p = 0.0014) and A. polyphaga (p = 0.0082) and that D. discoideum has a significantly lower infection intensity than A. lenticulata (p = 0.0455). These findings demonstrate genus-level differences in infection intensity. Infection intensity frequencies followed a strong negative binomial distribution (Figure 3). Each ameba species had several high-intensity outliers ranging up to a maximum of 84 intracellular bacteria observed in 1 A. lenticulata ameba (Figure 3).
Y. pestis resides in digestive and central vacuoles of both D. discoideum and A. castellanii amebae. Green fluorescent protein expressed by intracellular Y. pestis co-localizes with ameba vacuoles (Figure 2). TEM micrographs depict intracellular Y. pestis maintaining cellular shape and apparent membrane integrity inside A. castellanii ameba for <24 h postinfection (Figure 4). Ultrastructural analysis of TEM images reveals Y. pestis persistence within the niche of a tight-fitting vacuolar membrane visually similar to Yersinia-containing vacuoles (YCVs) observed in infected macrophages (27).
Y. pestis can survive inside D. discoideum amebae for >48 hours, but we found genus-level differences in intraameba survival of Y. pestis (Table). The bacterium did not survive beyond 24 h postinfection in A. castellanii, A. lenticulata, A. polyphaga, or V. vermiformis. However, Y. pestis co-cultured with D. discoideum exhibited consistent intracellular survival for >48 h postinfection under variable treatment conditions (Table; Figure 5). Y. pestis exhibited significantly higher survival/recoverability when co-cultured with amebae as compared to Y. pestis monoculture controls (p<0.001). Y. pestis monoculture controls yielded a mean of 17 CFUs after 1 h of gentamicin exposure with no recoverable control bacteria across all other treatments. Conversely, E. coli bacteria did not significantly persist under any treatment conditions when co-cultured with ameba (p<0.001). Uninfected ameba control lysates consistently yielded zero bacteria across all ameba species and treatments (data not shown). All Y. pestis co-cultures exposed to antibiotics had supernatants free of extracellular bacteria. Y. pestis had no cytopathic effect on any of the tested ameba species as verified by comparing ameba abundance between co-culture treatments and ameba monoculture controls (data not shown).
Y. pestis replicates intracellularly in D. discoideum amebae for >48 hours postinfection (Table; Figure 5). In D. discoideum, the abundance of viable intracellular Y. pestis was significantly greater at each successive time point (24 and 48 h postinfection) after 1 h of antibiotic drug exposure (p = 0.01 and p = 0.002, respectively). Additionally, the abundances of viable Y. pestis in D. discoideum at 24 and 48 h postinfection were significantly greater than immediately after the 4-h antibiotic treatment (p = 0.008 and p = 0.001, respectively). After 48 h postinfection, viable intracellular Y. pestis was only recovered from D. discoideum treatments. Because the data did not meet standard ANOVA assumptions of normality despite transformation attempts, we used a nonparametric Kruskal-Wallis rank-sum test to compare treatment means by species. Results indicated that the increased abundance of Y. pestis in D. discoideum was significant compared with all other species at 48 h postinfection (p<0.001).
We demonstrate that Y. pestis (CO-92) can survive and replicate intracellularly within the social, heterogamous ameba D. discoideum, whereas Y. pestis is only transiently resistant to 4 species of free-living and cyst-forming amebae (A. castellanii, A. lenticulata, A. polyphaga, and V. vermiformis). Relative to E. coli controls, Y. pestis demonstrated significantly increased survival and replication within amebae despite the 4 cyst-forming amebae successfully killing the bacteria by using unidentified mechanisms.
Amebae cultured from soil in prairie dog colonies with active plague epizootics confirm that ameba species used in our experiments co-occur spatially and temporally with Y. pestis under natural conditions. Interactions between amebae and Y. pestis could select for increasingly ameba-resistant phenotypes, considering the transient resistance already observed in 4 cyst-forming ameba species. Other research has demonstrated the potential for amebae to affect pathogen transmission mode, alter virulence, and act as training grounds for intracellular pathogens by selecting for traits enabling macrophage invasion or avoidance (17,38).
Genus-level differences in ameba infection intensity and infection prevalence confirm that various ameba species have greater reservoir potential than others. In accordance with super-spreader theories, a minority of individual ameba harboring atypically high numbers of intracellular bacteria may be disproportionately causative for pathogen maintenance and re-emergence.
We observed a shorter duration of survival for Y. pestis in A. castellanii compared with prior experiments (24 hs vs. 5 d in 13), likely from differing co-culture conditions and ameba strains. Incubation temperatures differed between this and prior experiments (28°C vs. 4°C and 25°C in 13). Many Y. pestis virulence factors are temperature regulated and may differentially facilitate cellular invasion, inhibition of the phagolysosomal pathway, and intracellular persistence (1,17,38). Additionally, A. castellanii (ATCC 30234) used in this study was originally derived from yeast cultures in London in 1930, whereas A. castellanii (ATCC 30010), used by Benavides-Montaño et al. (13), was originally isolated from California soil in 1957 and enabled longer intracellular survival of Y. pestis. Intracellular survival may be affected by traits acquired by co-evolution between amebae and resistant bacteria in soil environments (17).
In macrophages, Y. pestis recruits host Rab1b protein to the phagosome, resulting in inhibition of phagosome acidification and disruption of the remaining phagolysosomal metabolic pathway (26,27,39–41). Y. pestis then establishes a replicative niche within the YCV, characterized by a tight-fitting vacuole that expands commensurately with bacterial replication (27). Examination of TEM micrographs shows that intracellular bacteria are localized within form-fitting vacuolar membranes, similar to the YCVs found in macrophages (Figure 4).
The successful intracellular survival of Y. pestis in D. discoideum for >48 h demonstrates that Y. pestis is an ameba-resistant bacterium. This classification supports the potential for D. discoideum or related ameba species to be environmental reservoirs of plague. Intracellular survival of the observed duration is consequential given that typical interactions between bacteria and phagocytic cells result in bacterial death in <40 min (27). Most phagocytosed bacteria cannot survive digestive processes characteristic of phagocytic cells including phagolysosome fusion and acidification, or the subsequent recruitment of endosomal lytic factors (26,27,41). Ongoing research assesses the maintenance of viable Y. pestis through the entire D. discoideum life cycle, including transmissible dormant spores.
Ameba-resistant pathogens often replicate in vacuoles before escaping into the cytosol or outside of the phagocytic cell entirely. In addition to viable intracellular persistence, we observed active intracellular replication of Y. pestis (CO-92) in D. discoideum (Figure 5) and possible, but unconfirmed, replication of Y. pestis (CO92 pgm+, pCD1, pGFPuv, amp+) in A. castellanii (Figure 4, panel A). Analysis of TEM micrographs proved inconclusive for identifying the bacterial division septum; thus, only D. discoideum has conclusively demonstrated intracellular replication of Y. pestis. Y. pestis CFUs recovered from within D. discoideum increased significantly (p = 0.001–0.01; Figure 5) in successive postinfection time points across both antibiotic treatment conditions (except in 1 instance where Y. pestis increased nonsignificantly [p = 0.1624; Figure 5]). The consistent absence of extracellular bacteria in all D. discoideum co-cultures indicates resistance to digestion and the exploitation of an intraameba replicative niche.
Intracellular replication of Y. pestis in macrophages occurs within YCVs, and the formation of YCVs requires metabolic pathway inhibition by recruitment of Rab1b GTPases. Orthologous mechanisms are likely the cause for observed Y. pestis replication and survival within amebae. We searched for macrophage Rab1b by using BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi) against full genome sequences of each study ameba species and located homologous genetic sequences (99.8% similarity) within A. castellanii and D. discoideum (GenBank accession nos. XM_004347056.1 and XM_637217.1, respectively [42,43]). Future research should attempt to establish whether these ameba sequences are functionally orthologous to those identified in macrophages and whether the presence of particular host GTPases is diagnostic of ameba permissiveness to intracellular bacteria.
Results of this study support the reservoir potential of environmental ameba but do not definitively prove that this mechanism occurs in situ. Further research is necessary to determine if the maximum duration of intraameba Y. pestis survival corresponds with the durations of cryptic interepizootic persistence that are characteristic of plague dynamics. Increasing evidence for dormant or viable but nonculturable forms of Y. pestis may provide explanations underlying hypothesized multiyear survival in ameba spores or cysts (12,25,44–46). Outcomes of this research prompt questions regarding evolutionary selection imposed by amebae on environmental pathogens and applications of the ameba reservoir model for other cryptic environmental pathogens. Further research into ameba-mediated pathogenesis and persistence will offer practical insights for public health, conservation, agricultural management, and biodefense.
In conclusion, the mechanisms underlying plague re-emergence following dormancy have eluded researchers for centuries (1,11). Plague persistence within soilborne microorganisms has been hypothesized as an elusive maintenance mechanism (6,11,12,25). We demonstrated spatiotemporal co-occurrence of plague bacterium and various ameba species during an active plague epizootic. Further, we observed the persistence of viable and replicative Y. pestis in D. discoideum amebae for ˃48 hours postinfection and persistence of Y. pestis in 4 cyst-forming ameba species for <24 hours postinfection, whereas ameba-susceptible control bacteria were eliminated by amebae in <1 hour. Thus, Y. pestis are respectively ameba-resistant and transiently ameba-resistant under the tested infection conditions. Y. pestis bacteria resided within ameba structures that were visually analogous to YCVs observed in infected macrophages. These results encourage research into the eco-evolutionary interactions between pathogenic bacteria, amebae, and host immune factors. The reservoir potential of amebae and their shared infection-permissiveness with phagocytic macrophages show promise in explaining the cryptic properties underlying interepizootic plague transmission and persistence.
Mr. Markman is a PhD candidate at Colorado State University in the Department of Biology and is a concurrent research fellow for the National Defense, Science, and Engineering Graduate program of the US Department of Defense and the Vice President for Research program of Colorado State University. He collaborates with the CDC in Fort Collins, CO while researching cryptic bacterial persistence mechanisms that facilitate epidemics.
Acknowledgments
We thank K.L. Gage and S.W. Bearden for their aid in procuring bacterial isolates and constructive feedback on experimental design. We also thank A. Sanchez-Hidalgo for her assistance with microscopy.
Research was supported in part by a One Health Catalyst grant from the Office of the Vice President of Research at Colorado State University. D.W.M. was supported by National Science Foundation Integrative Graduate Education and Research Traineeship grant no. DGE-0966346 “I-WATER: Integrated Water, Atmosphere, Ecosystems Education and Research Program” to Colorado State University.
References
- Perry RD, Fetherston JD. Yersinia pestis—etiologic agent of plague. Clin Microbiol Rev. 1997;10:35–66.PubMedGoogle Scholar
- Bertherat E; World Health Organization. Plague around the world, 2010–2015. Wkly Epidemiol Rec. 2016;91:89–93.PubMedGoogle Scholar
- Webb CT, Brooks CP, Gage KL, Antolin MF. Classic flea-borne transmission does not drive plague epizootics in prairie dogs. Proc Natl Acad Sci U S A. 2006;103:6236–41. DOIPubMedGoogle Scholar
- Girard JM, Wagner DM, Vogler AJ, Keys C, Allender CJ, Drickamer LC, et al. Differential plague-transmission dynamics determine Yersinia pestis population genetic structure on local, regional, and global scales. Proc Natl Acad Sci U S A. 2004;101:8408–13. DOIPubMedGoogle Scholar
- Snäll T, O’Hara RB, Ray C, Collinge SK. Climate-driven spatial dynamics of plague among prairie dog colonies. Am Nat. 2008;171:238–48. DOIPubMedGoogle Scholar
- Eisen RJ, Gage KL. Adaptive strategies of Yersinia pestis to persist during inter-epizootic and epizootic periods. Vet Res. 2009;40:01.
- Gibbons HS, Krepps MD, Ouellette G, Karavis M, Onischuk L, Leonard P, et al. Comparative genomics of 2009 seasonal plague (Yersinia pestis) in New Mexico. PLoS One. 2012;7:e31604. DOIPubMedGoogle Scholar
- Lowell JL, Antolin MF, Andersen GL, Hu P, Stokowski RP, Gage KL. Single-nucleotide polymorphisms reveal spatial diversity among clones of Yersinia pestis during plague outbreaks in Colorado and the western United States. Vector Borne Zoonotic Dis. 2015;15:291–302. DOIPubMedGoogle Scholar
- Salkeld DJ, Stapp P, Tripp DW, Gage KL, Lowell J, Webb CT, et al. Ecological traits driving the outbreaks and emergence of zoonotic pathogens. Bioscience. 2016;66:118–29. DOIGoogle Scholar
- Pavlovsky EN. Natural nidality of transmissible diseases, with special reference to the landscape epidemiology of zooanthroponoses. Urbana (IL): University of Illinois Press; 1966.
- Gage KL, Kosoy MY. Natural history of plague: perspectives from more than a century of research. Annu Rev Entomol. 2005;50:505–28. DOIPubMedGoogle Scholar
- Ayyadurai S, Houhamdi L, Lepidi H, Nappez C, Raoult D, Drancourt M. Long-term persistence of virulent Yersinia pestis in soil. Microbiology. 2008;154:2865–71. DOIPubMedGoogle Scholar
- Benavides-Montaño JA, Vadyvaloo V. Yersinia pestis resists predation by Acanthamoeba castellanii and exhibits prolonged intracellular survival. Appl Environ Microbiol. 2017;83:e00593–17. DOIPubMedGoogle Scholar
- Greub G, Raoult D. Microorganisms resistant to free-living amoebae. Clin Microbiol Rev. 2004;17:413–33. DOIPubMedGoogle Scholar
- Hilbi H, Weber SS, Ragaz C, Nyfeler Y, Urwyler S. Environmental predators as models for bacterial pathogenesis. Environ Microbiol. 2007;9:563–75. DOIPubMedGoogle Scholar
- Bichai F, Payment P, Barbeau B. Protection of waterborne pathogens by higher organisms in drinking water: a review. Can J Microbiol. 2008;54:509–24. DOIPubMedGoogle Scholar
- Salah IB, Ghigo E, Drancourt M. Free-living amoebae, a training field for macrophage resistance of mycobacteria. Clin Microbiol Infect. 2009;15:894–905. DOIPubMedGoogle Scholar
- Thomas V, McDonnell G, Denyer SP, Maillard J-Y. Free-living amoebae and their intracellular pathogenic microorganisms: risks for water quality. FEMS Microbiol Rev. 2010;34:231–59. DOIPubMedGoogle Scholar
- Wheat WH, Casali AL, Thomas V, Spencer JS, Lahiri R, Williams DL, et al. Long-term survival and virulence of Mycobacterium leprae in amoebal cysts. PLoS Negl Trop Dis. 2014;8:e3405. DOIPubMedGoogle Scholar
- Pujol C, Bliska JB. The ability to replicate in macrophages is conserved between Yersinia pestis and Yersinia pseudotuberculosis. Infect Immun. 2003;71:5892–9. DOIPubMedGoogle Scholar
- Lambrecht E, Baré J, Chavatte N, Bert W, Sabbe K, Houf K. Protozoan cysts act as a survival niche and protective shelter for foodborne pathogenic bacteria. Appl Environ Microbiol. 2015;81:5604–12. DOIPubMedGoogle Scholar
- Santos-Montañez J, Benavides-Montaño JA, Hinz AK, Vadyvaloo V. Yersinia pseudotuberculosis IP32953 survives and replicates in trophozoites and persists in cysts of Acanthamoeba castellanii. FEMS Microbiol Lett. 2015;362:fnv091. DOIPubMedGoogle Scholar
- Savage LT, Reich RM, Hartley LM, Stapp P, Antolin MF. Climate, soils, and connectivity predict plague epizootics in black-tailed prairie dogs (Cynomys ludovicianus). Ecol Appl. 2011;21:2933–43. DOIGoogle Scholar
- Collinge SK, Johnson WC, Ray C, Matchett R, Grensten J, Cully JF Jr, et al. Landscape structure and plague occurrence in black-tailed prairie dogs on grasslands of the western USA. Landsc Ecol. 2005;20:941–55. DOIGoogle Scholar
- Easterday WR, Kausrud KL, Star B, Heier L, Haley BJ, Ageyev V, et al. An additional step in the transmission of Yersinia pestis? ISME J. 2012;6:231–6. DOIPubMedGoogle Scholar
- Ke Y, Chen Z, Yang R. Yersinia pestis: mechanisms of entry into and resistance to the host cell. Front Cell Infect Microbiol. 2013;3:106 . DOIPubMedGoogle Scholar
- Connor MG, Pulsifer AR, Price CT, Abu Kwaik Y, Lawrenz MB. Yersinia pestis requires host Rab1b for survival in macrophages. PLoS Pathog. 2015;11:e1005241. DOIPubMedGoogle Scholar
- Nikul’shin SV, Onatskaia TG, Lukanina LM, Bondarenko AI. [Associations of the soil amoeba Hartmannella rhysodes with the bacterial causative agents of plague and pseudotuberculosis in an experiment] [in Russian]. Zh Mikrobiol Epidemiol Immunobiol. 1992; (
9-10 ):2–5.PubMedGoogle Scholar - Pushkareva VI. [Experimental evaluation of interaction between Yersinia pestis and soil infusoria and possibility of prolonged preservation of bacteria in the protozoan oocysts] [in Russian]. Zh Mikrobiol Epidemiol Immunobiol. 2003; (
4 ):40–4.PubMedGoogle Scholar - Barker J, Brown MR. Trojan horses of the microbial world: protozoa and the survival of bacterial pathogens in the environment. Microbiology. 1994;140:1253–9. DOIPubMedGoogle Scholar
- Link VB. A history of plague in United States of America. Public Health Monogr. 1955;26:1–120.PubMedGoogle Scholar
- Lagkouvardos I, Shen J, Horn M. Improved axenization method reveals complexity of symbiotic associations between bacteria and acanthamoebae. Environ Microbiol Rep. 2014;6:383–8. DOIPubMedGoogle Scholar
- Charette SJ, Cosson P. Preparation of genomic DNA from Dictyostelium discoideum for PCR analysis. Biotechniques. 2004;36:574–5.PubMedGoogle Scholar
- Le Calvez T, Trouilhé M-C, Humeau P, Moletta-Denat M, Frère J, Héchard Y. Detection of free-living amoebae by using multiplex quantitative PCR. Mol Cell Probes. 2012;26:116–20. DOIPubMedGoogle Scholar
- Schuster FL. Cultivation of pathogenic and opportunistic free-living amebas. Clin Microbiol Rev. 2002;15:342–54. DOIPubMedGoogle Scholar
- Thomas V, Herrera-Rimann K, Blanc DS, Greub G. Biodiversity of amoebae and amoeba-resisting bacteria in a hospital water network. Appl Environ Microbiol. 2006;72:2428–38. DOIPubMedGoogle Scholar
- Fey P, Dodson R, Basu S, Chisholm R. One stop shop for everything Dictyostelium: dictyBase and the dicty stock center in 2012. In: Eichinger L, Rivero F, editors. Dictyostelium discoideum protocols. Methods in molecular biology (methods and protocols), vol 983. Totowa, NJ: Humana Press; 2013. p. 59–92.
- Molmeret M, Horn M, Wagner M, Santic M, Abu Kwaik Y. Amoebae as training grounds for intracellular bacterial pathogens. Appl Environ Microbiol. 2005;71:20–8. DOIPubMedGoogle Scholar
- Straley SC, Perry RD. Environmental modulation of gene expression and pathogenesis in Yersinia. Trends Microbiol. 1995;3:310–7. DOIPubMedGoogle Scholar
- Grabenstein JP, Fukuto HS, Palmer LE, Bliska JB. Characterization of phagosome trafficking and identification of PhoP-regulated genes important for survival of Yersinia pestis in macrophages. Infect Immun. 2006;74:3727–41. DOIPubMedGoogle Scholar
- Pujol C, Klein KA, Romanov GA, Palmer LE, Cirota C, Zhao Z, et al. Yersinia pestis can reside in autophagosomes and avoid xenophagy in murine macrophages by preventing vacuole acidification. Infect Immun. 2009;77:2251–61. DOIPubMedGoogle Scholar
- Clarke M, Lohan AJ, Liu B, Lagkouvardos I, Roy S, Zafar N, et al. Genome of Acanthamoeba castellanii highlights extensive lateral gene transfer and early evolution of tyrosine kinase signaling. Genome Biol. 2013;14:R11. DOIPubMedGoogle Scholar
- Eichinger L, Pachebat JA, Glöckner G, Rajandream M-A, Sucgang R, Berriman M, et al. The genome of the social amoeba Dictyostelium discoideum. Nature. 2005;435:43–57. DOIPubMedGoogle Scholar
- Buzoleva LS, Sidorenko ML. [Influence of gaseous metabolites of soil bacteria on the multiplication of Listeria monocytogenes and Yersinia pseudotuberculosis.] [in Russian]. Zh Mikrobiol Epidemiol Immunobiol. 2005; (
2 ):7–11.PubMedGoogle Scholar - Somova LM, Buzoleva LS, Isachenko AS, Somov GP. [Adaptive ultrastructural changes in soil-resident Yersinia pseudotuberculosis bacteria] [in Russian]. Zh Mikrobiol Epidemiol Immunobiol. 2006; (
3 ):36–40.PubMedGoogle Scholar - Pawlowski DR, Metzger DJ, Raslawsky A, Howlett A, Siebert G, Karalus RJ, et al. Entry of Yersinia pestis into the viable but nonculturable state in a low-temperature tap water microcosm. PLoS One. 2011;6:e17585. DOIPubMedGoogle Scholar
Figures
Table
Cite This ArticleTable of Contents – Volume 24, Number 2—February 2018
| EID Search Options |
|---|
|
|
|
|
|
|


Please use the form below to submit correspondence to the authors or contact them at the following address:
David W. Markman, Colorado State University, Department of Biology, 1878 Campus Delivery, Fort Collins, CO 80523, USA
Top